 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c3_37540V3.1.p | ||||||||
| Common Name | PpCCA1b | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 960aa MW: 101296 Da PI: 6.0098 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.3 | 2.7e-16 | 57 | 101 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE++++++a k++G W++I +++g ++t+ q++s+ qk+
Pp3c3_37540V3.1.p 57 RERWTEEEHQKFLEALKLYGRA-WRRIEEHIG-TKTAVQIRSHAQKF 101
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.87E-16 | 51 | 107 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.436 | 52 | 106 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 1.7E-16 | 55 | 104 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.0E-13 | 56 | 104 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-13 | 57 | 100 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-9 | 57 | 97 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.05E-10 | 59 | 102 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 960 aa Download sequence Send to blast |
MIPMTPSSSA AGYREHEGAG EEGTYWPSVS PGVPLSTSGD EGASTKVRKP YTITKQRERW 60 TEEEHQKFLE ALKLYGRAWR RIEEHIGTKT AVQIRSHAQK FFSKIERDVT AGQGTETGVA 120 QVIDIPPPRP KRKPTHPYPR KAGRSFGKTS EDECPLAAAG SIVSSSGVST ANISEACLKE 180 GVWDQDDTGV AGHQDAAKAK KDADTAHSWG VPKTVNSATS GSPTARPVNV ALNGGLPLNN 240 ISPVPIPGFP AMPPWNRLFG GAMNPAMMNP AMLNSAMMNP QGMLMPWATA PNVQGLENGG 300 AVSAMAATIA AASAWWAMQG GMPPGVMHPA LGAMYLGAAP PLAIPTPYIF SQCVPQEITV 360 AAAPCADGNE VPTAQVVDDG QEVLSNNNHS RSSAMRRAER AQRVKELKAR RMGSGNHLLS 420 VSSDICASTS GGSGTNLTTQ LTAGGLTSES AVNSLSKGSS LPADLSKVKH VLNGLIRKDR 480 EAPVDGCSAD DGVDNRKRLR GSRPSDDTAM PDGEGYHSTS GLGREAPGSM DSHLEKVRSV 540 SGSNSIPGKF TKVDVERLAC DEKTAPTQAS CGSDVSEGAG VAPIDGDGDG DASGGGPSSN 600 SSACGNVPGG NERSGSSDGS SEGEATCNVD RQREAGKNGT SSSDPEAEEE ADEKVGRAAE 660 QFTFNEFLPV SKDGDKHEKP ETSSRLPRTV SIDGEQRKEV TERGQIAFQA LFKRDTLPRT 720 FSPPPGLALG TGTKEAAKQQ PSVSTMSRSL SANAITCDSL ELRTSQRNHK DQDSAFNTWN 780 HLRGTSSASK VDRCESGISI PSSRPTKSLE DLRSSERRRG SRKGRSHTGA NNWLQLWTDT 840 SSLDKDEDAD EDDEEDESAD SGADDGCKKQ QQKASFVCKL DMSISNQEVE LISDVEGTGA 900 SSLNISSTVR SQTLFLSVGT GGLKNAVSNV PTRTVKYSGV GFVPYQRAST RTEPLVDKL* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppa.16227 | 0.0 | gametophore| protonema | ||||
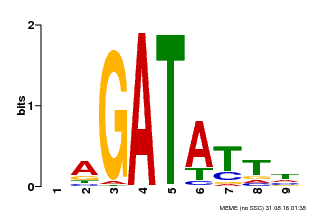
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00654 | PBM | Transfer from LOC_Os08g06110 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB458832 | 0.0 | AB458832.1 Physcomitrella patens PpCCA1b mRNA for circadian clock-associated protein 1b, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024372041.1 | 0.0 | uncharacterized protein LOC112280611 | ||||
| TrEMBL | A0A2K1KXL0 | 0.0 | A0A2K1KXL0_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S96_165V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 8e-35 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c3_37540V3.1.p |



