 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c4_2590V3.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 411aa MW: 43659 Da PI: 7.1606 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 42 | 2.2e-13 | 44 | 88 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT++E++++++a +++ + Wk+I + +g +t q++s+ qky
Pp3c4_2590V3.2.p 44 RESWTEQEHDKFLEALQLFDRD-WKKIEAFVG-SKTVIQIRSHAQKY 88
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.42E-16 | 38 | 94 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.408 | 39 | 93 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-6 | 42 | 94 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.0E-18 | 42 | 91 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.3E-11 | 43 | 91 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.7E-11 | 44 | 88 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.18E-8 | 46 | 89 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 411 aa Download sequence Send to blast |
MNAASSEMLG VMVAGSTLSS IPPIPVSEEG SKKIRKPYTI TKSRESWTEQ EHDKFLEALQ 60 LFDRDWKKIE AFVGSKTVIQ IRSHAQKYFL KVQKNGTGEH VPPPRPKRKS VQPYPQKAPK 120 TAQVQVSDGL RTQAVQTESS YGTGSHKPPM TSTSPSISAW VQHSVSPNPS ISYDAPGIKS 180 EGEGVNLSAV RVPSNSISGS SPGGWPQHVL PASQVAPESC IRAAPDFAEV YKFIGSVFDP 240 GVSGHLRKLK EMSAIDRETV LLLMHNLSIN LASPDFEEHD AGEDSPSVSG QAGTLAPSTP 300 ILVSPRSGNS APTQHRASLL QAYADSTADA GTARRHSHDG GRVISASFTS CGVSPAPEVL 360 EPVSLGLPCT ALQPQSPVLT TKPESVYSIT SLPSLPAGLA GDESVDTWWT * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00560 | DAP | Transfer from AT5G52660 | Download |
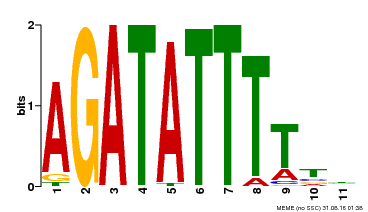
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024374226.1 | 0.0 | protein REVEILLE 3-like isoform X3 | ||||
| Swissprot | Q6R0H0 | 1e-83 | RVE3_ARATH; Protein REVEILLE 3 | ||||
| Swissprot | Q8H0W3 | 2e-83 | RVE6_ARATH; Protein REVEILLE 6 | ||||
| TrEMBL | A0A2K1KLY0 | 0.0 | A0A2K1KLY0_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S160_6V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G52660.2 | 1e-86 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c4_2590V3.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



