 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Pp3c8_16910V3.2.p | ||||||||
| Common Name | N3, PHYPADRAFT_166399 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Bryophyta; Bryophytina; Bryopsida; Funariidae; Funariales; Funariaceae; Physcomitrella
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 486aa MW: 53857.1 Da PI: 8.2257 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.4 | 1.2e-31 | 332 | 385 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH++Fv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
Pp3c8_16910V3.2.p 332 PRMRWTSTLHAHFVQAVELLGGHERATPKSVLELMNVKDLTLAHVKSHLQMYRT 385
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.65E-15 | 329 | 386 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.3E-28 | 330 | 385 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.1E-24 | 332 | 385 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.9E-7 | 333 | 384 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010229 | Biological Process | inflorescence development | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005618 | Cellular Component | cell wall | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0030003 | anatomy | protonema | ||||
| PO:0030018 | anatomy | gametophore | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 486 aa Download sequence Send to blast |
MQELSPDKSM SVSSSSNTHS GPDLALHISP PSSSSCDRRL KSSSQYSNLQ VVPDHVHGFD 60 LWKQPTKSMS CITDSESNVS SPAAEGYSPA YVNSMGASTV LCLANPPLDS QYVKRALPKS 120 TADEGMVIGR REEVQESVQE NGYICKGLAH PFLLDNWSES IRISPDRVSS PSDRIRKDLP 180 RTFSIYPNET QENFTRRLSE SSIDDLYSQQ RVFSRPNYTS SHPSPIHELS LGRWSDPGVH 240 PEALPSRDLP KFQNYYMVGM DSCHRFSHQY GLENGKNMLG MQASTCGGNL STLLGNYGAG 300 YNAKREGVYA NVTSFRSHFP PRSPAKRSIR APRMRWTSTL HAHFVQAVEL LGGHERATPK 360 SVLELMNVKD LTLAHVKSHL QMYRTVKTSD KSGPPSGSRD PSPTSSLDSR LTTEELLMSD 420 SMSQTGRSVH MSNSTMHNLE LGNYRSLGDS QDATVSMQNR VWLPENPQNF NQAQRGCWKF 480 GTLSC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 7e-17 | 333 | 387 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 7e-17 | 333 | 387 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 6e-17 | 333 | 387 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 6e-17 | 333 | 387 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 6e-17 | 333 | 387 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 6e-17 | 333 | 387 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 7e-17 | 333 | 387 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 7e-17 | 333 | 387 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 7e-17 | 333 | 387 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 7e-17 | 333 | 387 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 7e-17 | 333 | 387 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 7e-17 | 333 | 387 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppa.18248 | 0.0 | protonema | ||||
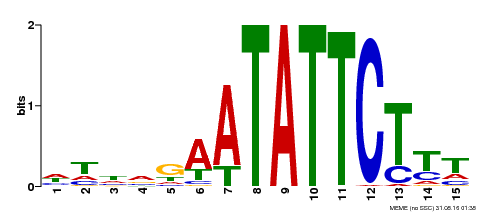
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024382995.1 | 0.0 | uncharacterized protein LOC112285891 | ||||
| TrEMBL | A0A2K1K7L3 | 0.0 | A0A2K1K7L3_PHYPA; Uncharacterized protein | ||||
| STRING | PP1S114_151V6.1 | 0.0 | (Physcomitrella patens) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32240.1 | 2e-36 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Pp3c8_16910V3.2.p |
| Entrez Gene | 5932621 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



