 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Prupe.1G019100.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 795aa MW: 87145.4 Da PI: 6.1335 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 49.8 | 7e-16 | 496 | 534 | 2 | 40 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
++k+CnCkkskClk+YCeCfaag++C e C+C++C+Nk
Prupe.1G019100.1.p 496 SCKRCNCKKSKCLKLYCECFAAGVYCIEPCSCQECFNKP 534
689**********************************96 PP
| |||||||
| 2 | TCR | 50.5 | 4.1e-16 | 582 | 620 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN
Prupe.1G019100.1.p 582 RHKRGCNCKKSSCLKKYCECYQGGVGCSISCRCEGCKNA 620
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 2.5E-17 | 495 | 536 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 37.28 | 496 | 622 | IPR005172 | CRC domain |
| Pfam | PF03638 | 1.6E-11 | 498 | 533 | IPR005172 | CRC domain |
| SMART | SM01114 | 4.4E-17 | 582 | 623 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 1.3E-11 | 584 | 620 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 795 aa Download sequence Send to blast |
MDTPERNQIG TPKAKFEDSP VFNYINSLSP IKPVKSVHIT QTFSSLSFAS LPSVFTSPHV 60 SAHKESRFLR RHNISDPSKP ESSLESGNKV SANEDAAQLY INSEELREDC VQGVSTGEDS 120 VEPSSEHSKF VIELPRNLKY DCGSPDCHPT TRCGTEAHCE LEVADLSAPL VPYVQKTSEE 180 GSSSDEAHLQ IICQTVQRKE GTGCDWESLI CDAADLLIFD SPNGTEAFKG LMQNSLDPVT 240 RFCTSLAPQL TQNDVNDEQN VQVLDMVGSG GQLETEDPAS QYGEASKLER TEQMEGHLNN 300 CMVSSQSEKE DNKVETPLQF NCKPAVLNLQ RGLRRRCLDF EMAGARRKSL DNVSNSSSNM 360 LSQSDEKIAT NDKQLVPMKP GGESSRCILP GIGLHLNALA KTSKDYKIIK CESLAYGRQL 420 SLPNSTADIH SPTGQGPGHE SFSSASSERD MDGTENGVQL AHDASQEPAF LANEEFNQNS 480 PKKKRRRFEH AGETESCKRC NCKKSKCLKL YCECFAAGVY CIEPCSCQEC FNKPIHEDTV 540 LATRKQIESR NPLAFAPKVI RSSDSVPELG QEESSKTPAS ARHKRGCNCK KSSCLKKYCE 600 CYQGGVGCSI SCRCEGCKNA FGRKDGSFIG TEAELDEEEA EACEKSVAEK HQQKIEIQKN 660 EEQRLDSALP TTPLRLSRQM VSLPFSSKNK PPRSSVFSIG GSSSGLYTSQ KLGQPNILRP 720 ESKFERHSQS VPEDEMPEIL QGDVSPSTGV KTASPNSKRV CPPNTDFGPS PGRRTGRKLI 780 LQSIPSFPSL TPQH* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 5e-17 | 498 | 619 | 12 | 120 | Protein lin-54 homolog |
| 5fd3_B | 5e-17 | 498 | 619 | 12 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 481 | 486 | KKKRRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
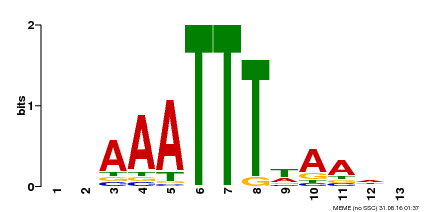
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Prupe.1G019100.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020410865.1 | 0.0 | CRC domain-containing protein TSO1 isoform X1 | ||||
| TrEMBL | A0A251QRE6 | 0.0 | A0A251QRE6_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008224266.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2619 | 33 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G14770.1 | 1e-142 | TESMIN/TSO1-like CXC 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Prupe.1G019100.1.p |



