 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Prupe.1G238200.3.p | ||||||||
| Common Name | PRUPE_ppa002679mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 616aa MW: 68903.1 Da PI: 5.897 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 69.1 | 7.3e-22 | 199 | 251 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++W+ +LH++Fv+av+q+ G + Pk+il+lm+v+ Lt+e+v+SHLQkYRl
Prupe.1G238200.3.p 199 KARVVWSVDLHQKFVKAVHQI-GFD-IGPKKILDLMNVPWLTRENVASHLQKYRL 251
68*******************.777.78**************************8 PP
| |||||||
| 2 | Response_reg | 81.4 | 2.9e-27 | 19 | 127 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHH CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalea 89
vl+vdD+p+ +++l+++l+k y ev+++ +++al+ll+ek+ +D+++ D++mp+mdG++ll++ e +lp+i+++ ge + + +
Prupe.1G238200.3.p 19 VLVVDDDPTWLKILEKMLKKCSY-EVTTCGLARDALNLLREKKdgYDIVVSDVNMPDMDGFKLLEHVGLEM-DLPVIMMSVDGETSRVMKG 107
89*********************.***************999999**********************6644.8****************** PP
HHTTESEEEESS--HHHHHH CS
Response_reg 90 lkaGakdflsKpfdpeelvk 109
++ Ga d+l Kp+ ++el++
Prupe.1G238200.3.p 108 VQHGACDYLLKPIRMKELRN 127
*****************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 2.1E-145 | 1 | 586 | IPR017053 | Response regulator B-type, plant |
| SuperFamily | SSF52172 | 6.4E-35 | 16 | 139 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 5.8E-44 | 16 | 141 | No hit | No description |
| SMART | SM00448 | 2.5E-32 | 17 | 129 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 43.134 | 18 | 133 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 3.5E-24 | 19 | 128 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 3.89E-30 | 20 | 133 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-25 | 197 | 256 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 5.38E-15 | 197 | 255 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-19 | 199 | 251 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 5.3E-8 | 201 | 250 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 616 aa Download sequence Send to blast |
MESSFSSPRN DSFPAGLRVL VVDDDPTWLK ILEKMLKKCS YEVTTCGLAR DALNLLREKK 60 DGYDIVVSDV NMPDMDGFKL LEHVGLEMDL PVIMMSVDGE TSRVMKGVQH GACDYLLKPI 120 RMKELRNIWQ HVFRKKIHEI RDIESHESIE GIQLIRNGSD QYDEGYFFSA EDLTSSRKRK 180 DVDNKYDDKD FSDCSSAKKA RVVWSVDLHQ KFVKAVHQIG FDIGPKKILD LMNVPWLTRE 240 NVASHLQKYR LYLSRLQKEN DIKSSFGGTK HSDYSSKDLQ GILSVQNSIN IQQNDVAHGS 300 YRFSGNNLVA QNADPKSNEG DIKGIVSEPV AEPKKGTTGN IPDSQKTRSS QMDFNSSYAE 360 TQKGLVGNVP DSQKIRSSQM RVNHSFASVE SEVNLTAFDS TIPATYSWNK IQLEKEHKPL 420 IQLNNGFSKP QLPGPQHHFQ ADQLQSIPSI SSRPSIPGGD VTGSSKSKPS YSEYTNHHGS 480 HVSPTISPTI SATDSFPGQT KSCVVDHQLS ETISTSTSNM KNQGINLSTD LESAQRNLIL 540 GSTSPFASLD DEFQICWYQG DCYGMNLELQ NVEFSEYTDP THVTEVPAHL YDALRFEYPC 600 DPSEYSVLDQ GLFIA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-15 | 195 | 256 | 1 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
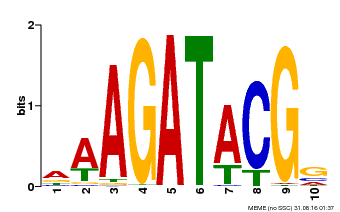
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Prupe.1G238200.3.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020409742.1 | 0.0 | two-component response regulator ARR11 | ||||
| TrEMBL | A0A251R2C3 | 0.0 | A0A251R2C3_PRUPE; Two-component response regulator | ||||
| STRING | EMJ28155 | 0.0 | (Prunus persica) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-136 | response regulator 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Prupe.1G238200.3.p |
| Entrez Gene | 18789818 |



