 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Prupe.1G290100.3.p | ||||||||
| Common Name | PRUPE_ppa014900mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 180aa MW: 19574.3 Da PI: 8.4964 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 145.8 | 4.5e-45 | 13 | 157 | 4 | 163 |
YABBY 4 fssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakasqllaaeshldeslkeelleelkveeenlksnvekeesast 94
++ se++Cyv+CnfC+t+lav +P++ l+ +vtv+CGhC++l ++ + a q + ++h l + +++ + +s+
Prupe.1G290100.3.p 13 NQPSEHLCYVRCNFCSTVLAVGLPFKRLLDTVTVKCGHCSNLSFLSTRPALQGQCLSDH---------PTSLTLQAGCCSEFR---KGQSS 91
5789*************************************987777777766666666.........222222222222222...22222 PP
YABBY 95 svsseklsenedeevprvppvirPPekrqrvPsaynrfikeeiqrikasnPdishreafsaaaknWahf 163
s ss + +++ ++p++p v++PPek+ r Psaynrf+keeiqrika+nP+i hreafsaaaknWa +
Prupe.1G290100.3.p 92 SSSS---PISSEPSSPKAPFVVKPPEKKHRLPSAYNRFMKEEIQRIKAANPEIPHREAFSAAAKNWARY 157
2222...234566789999************************************************76 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 3.5E-54 | 15 | 157 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 7.46E-9 | 101 | 155 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 3.1E-5 | 111 | 156 | IPR009071 | High mobility group box domain |
| CDD | cd01390 | 3.29E-6 | 118 | 154 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010254 | Biological Process | nectary development | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0048479 | Biological Process | style development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 180 aa Download sequence Send to blast |
MNLEEKVTMD LINQPSEHLC YVRCNFCSTV LAVGLPFKRL LDTVTVKCGH CSNLSFLSTR 60 PALQGQCLSD HPTSLTLQAG CCSEFRKGQS SSSSSPISSE PSSPKAPFVV KPPEKKHRLP 120 SAYNRFMKEE IQRIKAANPE IPHREAFSAA AKNWARYIPS PSAGSDSGSC SSTNNILQE* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Detected during flower and leaf development. Expression in the flower meristem in the early stage of flower development. When carpel primordia begin to form, specific and uniform expression in carpel primordia. Expression in the central region of the leaf plastochron 1 (P1) primordia. Detected up to P4 stage, hardly detected in the P5 leaves. {ECO:0000269|PubMed:14729915}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Regulates carpel specification in flower development. Severe or intermediate mutation in DL causes complete or partial homeotic conversion of carpels into stamens without affecting the identities of other floral organs. Interacts antagonistically with class B genes and controls floral meristem determinacy. Regulates midrib formation in leaves probably by inducing cell proliferation in the central region of the leaf. {ECO:0000269|PubMed:14729915}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00220 | DAP | Transfer from AT1G69180 | Download |
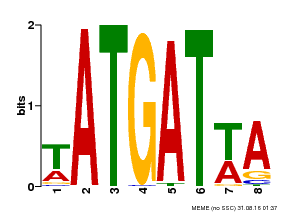
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Prupe.1G290100.3.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB627250 | 1e-118 | AB627250.1 Malus x domestica mRNA, microsatellite: MEST098, clone: FLW01_10_E08. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008243820.1 | 1e-130 | PREDICTED: protein CRABS CLAW isoform X2 | ||||
| Swissprot | Q76EJ0 | 8e-65 | YABDL_ORYSJ; Protein DROOPING LEAF | ||||
| TrEMBL | A0A251R4V9 | 1e-129 | A0A251R4V9_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008243820.1 | 1e-129 | (Prunus mume) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69180.1 | 3e-62 | YABBY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Prupe.1G290100.3.p |
| Entrez Gene | 18793592 |



