 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Prupe.1G393000.1.p | ||||||||
| Common Name | PRUPE_ppa008566mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 328aa MW: 36576.1 Da PI: 7.8542 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 90.2 | 1.6e-28 | 172 | 231 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
++Dgy+WrKYGqK+ ++++ pr+YY+C++a +Cpvkkkv++sae+p +++ tYegeHnh
Prupe.1G393000.1.p 172 VKDGYQWRKYGQKVTRDNPSPRAYYKCSFApSCPVKKKVQKSAENPCLLVATYEGEHNHM 231
58***************************99****************************5 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.8E-28 | 162 | 233 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 25.935 | 167 | 233 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.88E-24 | 169 | 233 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.8E-31 | 172 | 232 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.3E-23 | 173 | 230 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0002237 | Biological Process | response to molecule of bacterial origin | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050691 | Biological Process | regulation of defense response to virus by host | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 328 aa Download sequence Send to blast |
MDSEWVNTSL GLNVIPFHPA SDDQAPVKKQ QELEGDYAKV FEGHASVKQE AAASHVLTEE 60 LNRISLENKK LTEMLTALCE NYTNLQSHVK ELMTAKQQSS DQNDLATNFN KKRKPDSEDY 120 RNMIGLTSPG TNTETSSISD EEYPCKRPKE NMNLKISKVY VRTEASDTRL IVKDGYQWRK 180 YGQKVTRDNP SPRAYYKCSF APSCPVKKKV QKSAENPCLL VATYEGEHNH MHPETRAEVT 240 LIGSISPNQQ LSPLSPSMAK RTSPVPTFSC DKNNNLSPRE IEGAPPAFQQ FLVQQMASSL 300 TKDPNFASAL AAAISGRFSD HSRMGNW* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 3e-19 | 171 | 233 | 13 | 74 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00465 | DAP | Transfer from AT4G31800 | Download |
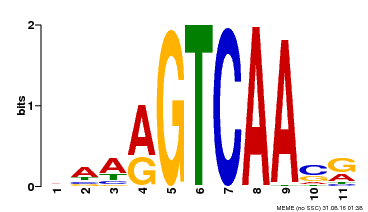
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Prupe.1G393000.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020411397.1 | 0.0 | WRKY transcription factor 18 | ||||
| TrEMBL | A0A251RCV9 | 0.0 | A0A251RCV9_PRUPE; Uncharacterized protein | ||||
| STRING | EMJ23850 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF973 | 34 | 115 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31800.1 | 5e-75 | WRKY DNA-binding protein 18 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Prupe.1G393000.1.p |
| Entrez Gene | 18789733 |



