 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Prupe.1G424300.1.p | ||||||||
| Common Name | PRUPE_ppa009924mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 273aa MW: 28608.7 Da PI: 8.8785 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13 | 0.00032 | 107 | 129 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
ykC+ C+k F++ L H +H
Prupe.1G424300.1.p 107 YKCSVCNKAFPSYQALGGHKASH 129
9***********99999998887 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 4.25E-9 | 107 | 129 | No hit | No description |
| SMART | SM00355 | 0.0057 | 107 | 129 | IPR015880 | Zinc finger, C2H2-like |
| Pfam | PF13912 | 6.4E-14 | 107 | 131 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 9.951 | 107 | 129 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 109 | 129 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 4.25E-9 | 157 | 184 | No hit | No description |
| Pfam | PF13912 | 2.3E-13 | 161 | 186 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.19 | 162 | 184 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.411 | 162 | 184 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 164 | 184 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006979 | Biological Process | response to oxidative stress | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009644 | Biological Process | response to high light intensity | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010117 | Biological Process | photoprotection | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0015979 | Biological Process | photosynthesis | ||||
| GO:0035264 | Biological Process | multicellular organism growth | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 273 aa Download sequence Send to blast |
MALQALNSPT TAGASPFRFE EAASKINYVE PWTKRKRSKR PRLDSPSTEE EYLALCLIML 60 ARGGSGSTSA ATTATNSAAS HHMITQSPSN ASHQEPSTSA QPPKLSYKCS VCNKAFPSYQ 120 ALGGHKASHR KGAAGSAAEG PSTSSNTTTS GTTTVTASGR THECSICHKS FPTGQALGGH 180 KRCHYEGGVG ASTATATATT TSAVTSSEGV GSTSHQAVSH GHRETFDLNM PALPEFSRDF 240 FLCGEDEVES PHPAKKPRIL TIMKPKQEIS QN* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppe.6214 | 0.0 | fruit| leaf | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00162 | DAP | Transfer from AT1G27730 | Download |
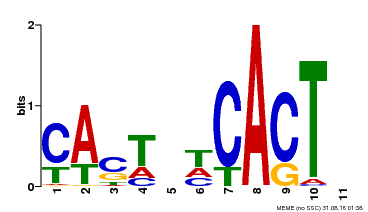
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Prupe.1G424300.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007222874.1 | 0.0 | zinc finger protein ZAT10 | ||||
| TrEMBL | M5XRA3 | 0.0 | M5XRA3_PRUPE; Uncharacterized protein | ||||
| STRING | EMJ24073 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1327 | 32 | 91 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G27730.1 | 2e-48 | salt tolerance zinc finger | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Prupe.1G424300.1.p |
| Entrez Gene | 18792372 |



