 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Prupe.1G551600.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 315aa MW: 36420.2 Da PI: 8.5028 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16.3 | 2.8e-05 | 36 | 60 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
y C+ dC+ s++rk++L+rH+ H
Prupe.1G551600.2.p 36 YICSvdDCHSSYRRKDHLNRHLLQH 60
89*******************9887 PP
| |||||||
| 2 | zf-C2H2 | 14.2 | 0.00013 | 65 | 90 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kCp +C++ F + n++rH + H
Prupe.1G551600.2.p 65 FKCPieNCNREFGFQGNMRRHVNElH 90
89*******************99888 PP
| |||||||
| 3 | zf-C2H2 | 15.9 | 3.8e-05 | 105 | 129 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ Cgk+F+ s L++H +H
Prupe.1G551600.2.p 105 HVCKeiGCGKVFRFASRLRKHEDSH 129
79*******************9888 PP
| |||||||
| 4 | zf-C2H2 | 15.7 | 4.3e-05 | 196 | 220 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC C +F++ksnL++H + H
Prupe.1G551600.2.p 196 KCDykGCLHTFTTKSNLTKHVKAvH 220
688889**************99877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 2.8E-9 | 32 | 62 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 5.67E-6 | 34 | 62 | No hit | No description |
| SMART | SM00355 | 0.0092 | 36 | 60 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.66 | 36 | 65 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 38 | 60 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.5E-5 | 63 | 86 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 7.66E-6 | 63 | 91 | No hit | No description |
| PROSITE profile | PS50157 | 11.718 | 65 | 95 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.014 | 65 | 90 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 67 | 90 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.9E-7 | 100 | 129 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.201 | 105 | 134 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.031 | 105 | 129 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 107 | 129 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 8.6 | 137 | 162 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 139 | 162 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 26 | 165 | 186 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 2.98E-6 | 180 | 232 | No hit | No description |
| PROSITE profile | PS50157 | 11.759 | 195 | 225 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.003 | 195 | 220 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.8E-6 | 196 | 220 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 197 | 220 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.0E-5 | 221 | 249 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 7.6 | 226 | 252 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 228 | 252 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 315 aa Download sequence Send to blast |
MRDGYDLNGL VPGGRPPVKL HQVNLNHALF GLRRPYICSV DDCHSSYRRK DHLNRHLLQH 60 QGKLFKCPIE NCNREFGFQG NMRRHVNELH SEDSPSTNGG AQKQHVCKEI GCGKVFRFAS 120 RLRKHEDSHV KLDTVEAFCS EPGCMKHFTN EQCLQAHIHS CHQHTTCEIC GTKQLRNNIK 180 RHLLTHEDKH SIERIKCDYK GCLHTFTTKS NLTKHVKAVH LEHKPFVCSF SGCGLRFAYK 240 HVRDNHEKTG CHVYTQGDFE EADEQFRSRP RGGRKRECPD IPMLVRKRVT PPNQLGQECE 300 YISWLHSQED NDEQ* |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Both isoforms 1 and 2 transcripts are expressed in seedlings and both TFIIIA and TFIIIA-C are present at similar ratios in 10-day-old seedlings (at protein level). In correlation with the reorganization of 5S rDNA chromatin to a mature state, full-length isoform 1 protein TFIIIA with transcriptional activity accumulates and permits de novo transcription of 5S rRNA. Isoforms 1 and 2 transcripts accumulate in various tissues of the reproductive phase, including flowers and siliques, but only isoform 2 is present in mature seeds. In flowers, both TFIIIA and TFIIIA-C are present at similar ratios (at protein level). Very low amounts of TFIIIA are found in extracts from fresh siliques compared to TFIIIA-C. The amount of full-length TFIIIA protein progressively increases from fresh siliques to seeds concomitant with lower proportions of the shorter TFIIIA-C protein (at protein level) thus leading to 5S rRNA accumulation in the seed. {ECO:0000269|PubMed:22353599}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in seedlings, flowers, siliques and seeds. {ECO:0000269|PubMed:22353599}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
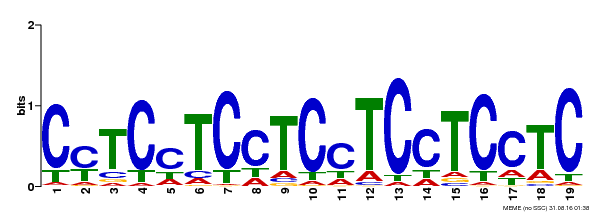
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Prupe.1G551600.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020410711.1 | 0.0 | transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 1e-113 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A251RI77 | 0.0 | A0A251RI77_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008219031.1 | 0.0 | (Prunus mume) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-115 | transcription factor IIIA | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Prupe.1G551600.2.p |



