 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Prupe.6G193400.1.p | ||||||||
| Common Name | PRUPE_ppa008318mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 338aa MW: 38048.2 Da PI: 4.9818 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.3 | 6.1e-19 | 58 | 111 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe ++++ e++ +LA++lgL+ rqV vWFqNrRa++k
Prupe.6G193400.1.p 58 KKRRLSVEQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWK 111
56689************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 130.6 | 6.4e-42 | 57 | 149 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrls eqvk+LE++Fe e+kLeperKv+la+eLglqprqvavWFqnrRAR+ktkqlE+d+ +Lk++yd+lk + ++L++e+e+L +e+
Prupe.6G193400.1.p 57 EKKRRLSVEQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWKTKQLERDFGVLKANYDSLKLNYDNLQHENEALVKEI 147
69***************************************************************************************99 PP
HD-ZIP_I/II 92 ke 93
k+
Prupe.6G193400.1.p 148 KQ 149
87 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.28E-19 | 45 | 115 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.572 | 53 | 113 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 6.1E-18 | 56 | 117 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 4.0E-16 | 58 | 111 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 5.22E-16 | 58 | 114 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-19 | 60 | 118 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 9.9E-6 | 84 | 93 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 88 | 111 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 9.9E-6 | 93 | 109 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.2E-17 | 113 | 155 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 338 aa Download sequence Send to blast |
MKRLGSSDSL GAMISICPST AEEQSPRNNH VYRRDFQSML DGLDEEGCVE EGGHVSEKKR 60 RLSVEQVKAL EKNFEVENKL EPERKVKLAQ ELGLQPRQVA VWFQNRRARW KTKQLERDFG 120 VLKANYDSLK LNYDNLQHEN EALVKEIKQL KSKLQEENTE SNNLSVKEEQ MVAKDQSNYK 180 VVDHELSKSP PPPPLGSSVP ATESKELNFE SFNNTNNGAV GLEAVSLFPD FKDGSSDSDS 240 SAILNEDNSP NLTISSSGML QNHQLMKSPA STSLKFNCCS SSSPSSSSMN CFQFQKTYHP 300 QFVKIEEHNF FSSEEACSFF SDEQAPTLQW CCPDQWN* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 105 | 113 | RRARWKTKQ |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Ppe.1420 | 0.0 | fruit | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
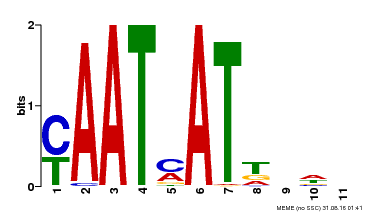
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Prupe.6G193400.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB621908 | 4e-92 | AB621908.1 Pyrus pyrifolia mRNA, microsatellite: TsuENH184. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007205507.1 | 0.0 | homeobox-leucine zipper protein ATHB-6 isoform X1 | ||||
| TrEMBL | M5W009 | 0.0 | M5W009_PRUPE; Uncharacterized protein | ||||
| STRING | EMJ06706 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1868 | 34 | 89 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22430.1 | 5e-67 | homeobox protein 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Prupe.6G193400.1.p |
| Entrez Gene | 18772872 |



