 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Prupe.6G271600.1.p | ||||||||
| Common Name | PRUPE_ppa009103mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 307aa MW: 34662.5 Da PI: 8.8072 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 26.1 | 1.9e-08 | 28 | 72 | 3 | 45 |
SS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwq 45
WT+eE +++ +a +++ ++ +W ++a++++ g+t +++ +
Prupe.6G271600.1.p 28 EWTAEENKQFENALALYDKDtpdRWFKVAARIP-GKTVGDVIKQYK 72
6********************************.****99988775 PP
| |||||||
| 2 | Myb_DNA-binding | 47.2 | 5.1e-15 | 135 | 179 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ +++ + k++G+g+W+ I+r + ++Rt+ q+ s+ qky
Prupe.6G271600.1.p 135 PWTEEEHRQFLMGLKKYGKGDWRNISRNFVTTRTPTQVASHAQKY 179
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 9.472 | 24 | 79 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.2E-10 | 25 | 77 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 4.67E-11 | 27 | 81 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 4.1E-7 | 28 | 72 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.18E-8 | 28 | 74 | No hit | No description |
| PROSITE profile | PS51294 | 20.486 | 128 | 184 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.94E-17 | 130 | 183 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.2E-17 | 131 | 183 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 5.7E-13 | 132 | 179 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.1E-13 | 132 | 182 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.0E-12 | 135 | 179 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.55E-11 | 135 | 180 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 307 aa Download sequence Send to blast |
MYRGIGILSP TSYLENSNWL FQESNGTEWT AEENKQFENA LALYDKDTPD RWFKVAARIP 60 GKTVGDVIKQ YKELEEDVSD IEAGLIPIPG YTSNSFTLEW VNNQGFDGLK QFYGVGGKRG 120 TSTRPADQER KKGVPWTEEE HRQFLMGLKK YGKGDWRNIS RNFVTTRTPT QVASHAQKYF 180 IRQLTGGKDK RRSSIHDITT VNLQDNKPPS PDSKSPPSSD HSSTVVQSQQ HQKVTGMSEQ 240 QFDWKSPNEG QPMVFNSANG STMGPFCGIS SYVPKLEEQN FLSGNLHGSQ LGHYNARFQM 300 QSMRYQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 2e-16 | 25 | 93 | 7 | 75 | RADIALIS |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Detected in the apical inflorescence meristem, in bract primordia arising in its periphery and in floral meristems produced in the axils of bracts (stages 0-3). From stage 3 to stage 8, detected in all floral organs irrespective of their dorsoventral positions. From stage 9, barely detectable in bracts, sepals, and stamens. In the corolla, however, expression was maintained and enhanced in some regions. Within ventral and lateral petals at stage 9, asymmetric pattern of expression with high levels of transcripts in the inner epidermis of the furrow and very reduced levels in the remaining cell layers. In the dorsal petals, from stage 9 onward, detected but with a more even distribution across cell layers than in the ventral petal. {ECO:0000269|PubMed:11937495}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00296 | DAP | Transfer from AT2G38090 | Download |
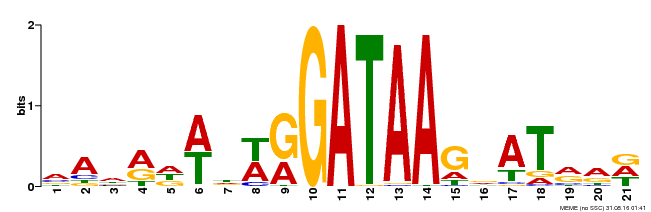
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Prupe.6G271600.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GQ246163 | 0.0 | GQ246163.1 Malus x domestica cultivar Jiangsu Fuji MYB transcription factor (MYB53) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007205616.1 | 0.0 | transcription factor DIVARICATA | ||||
| Swissprot | Q8S9H7 | 1e-120 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | M5WJ30 | 0.0 | M5WJ30_PRUPE; Uncharacterized protein | ||||
| STRING | EMJ06815 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF466 | 34 | 162 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38090.1 | 1e-121 | MYB family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Prupe.6G271600.1.p |
| Entrez Gene | 18772011 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



