 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC10120_p1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 725aa MW: 81347.1 Da PI: 6.3008 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 162 | 1.1e-50 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcy 99
+e +rwl+++ei+a+L n++ +++ ++ + pksg++++++rk++r+frkDG++wkkkkdgkt++E+he+LKvg+ e +++yYah+++nptf rrcy
RrC10120_p1 30 EEaYTRWLRPNEIHALLSNHNYFTINVKPVHLPKSGTILFFDRKMLRNFRKDGHNWKKKKDGKTIKEAHEHLKVGNEERIHVYYAHGDDNPTFVRRCY 127
55589********************************************************************************************* PP
CG-1 100 wlLeeelekivlvhylevk 118
wlL++++e+ivlvhy+e++
RrC10120_p1 128 WLLDKSQEHIVLVHYRETH 146
****************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 75.573 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.1E-73 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.6E-45 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.36E-14 | 368 | 454 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF12796 | 7.2E-7 | 554 | 634 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.4E-15 | 555 | 667 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.18E-15 | 563 | 670 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.80E-12 | 570 | 664 | No hit | No description |
| PROSITE profile | PS50297 | 13.761 | 572 | 637 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 2.7E-6 | 605 | 634 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.808 | 605 | 637 | IPR002110 | Ankyrin repeat |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 725 aa Download sequence Send to blast |
MAGVDSARLI GSEIHGFHTL QDLDIKTMLE EAYTRWLRPN EIHALLSNHN YFTINVKPVH 60 LPKSGTILFF DRKMLRNFRK DGHNWKKKKD GKTIKEAHEH LKVGNEERIH VYYAHGDDNP 120 TFVRRCYWLL DKSQEHIVLV HYRETHEVQA APATPGNSYS SSTTDHLSAK PLAEDNNSGV 180 RNPARSNSLV ARNHEISLHE INTLDWDELL VEADISNQPS PTEDDVLYFT EQLQTAPMGS 240 AQQGNHHAVY NGSTDIPSYL GLGDHVYQNN SPCGAREFSS QHLHCVVDPN QQTRDPSAAV 300 ANEQGDALLN NGYGSQESFG KWVNNFISDS PGSVDDPSLE AVYTPGQESS APPAVFHPQS 360 NIPEQIFNIT DVSPAWAYST EKTKILVTGF FHDSFQHFGR SNLFCICGEV RVPAEFLQMG 420 VYRCFLPPQS PGIVNLYLSA DGTKPISQLF SFEHRSVPVI EKVAPQEDQL YKWEEFEFQV 480 RLAHLLFTSS NKISVFSSKI SPDNLLEAKK LASRTSHLLN SWAYLMKSIQ ANEVPFDQAR 540 DHLFELTLKN RLKEWLLEKV IEDRNTKEYD SKGLGVIHLC AVLGYTWSIL LFSWANISLD 600 FRDKHGWTAL HWAAYYGREK MVAALLSAGA RPNLVTDPTK EYLGGCTAAD IAQQKGYEGL 660 AAFLAEKCLV AQFRDMKMAG NISGNLEGIK AETSANPGHS NEEEQSLKDT LGRRGGCTDP 720 GSFQR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
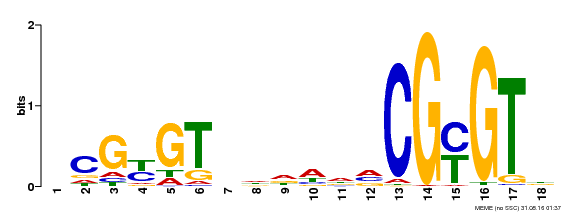
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY128295 | 0.0 | AY128295.1 Arabidopsis thaliana AT4g16150/dl4115w mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018450972.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A0D3DM50 | 0.0 | A0A0D3DM50_BRAOL; Uncharacterized protein | ||||
| STRING | Bo8g043280.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4021 | 27 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



