 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC10414_p1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 581aa MW: 65362 Da PI: 4.7859 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 154.9 | 3.6e-48 | 13 | 147 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..........kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkd 89
+pGfrFhPtdeelvv+yLk+k+ g+kl++ +vi vd+ykv+P +Lpk +k+++++w++F++r++ky++ r++r t++gyWkatgkd
RrC10414_p1 13 APGFRFHPTDEELVVYYLKRKICGRKLRI-NVIGVVDVYKVDPTELPKlnnlceaglsVLKSGDRQWFYFTPRNRKYPNAARSSRGTATGYWKATGKD 109
69***************************.99***************778999999987788999********************************* PP
NAM 90 kevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
+ ++ +++ vglkktLvfy+grap+ge+tdWvmhey++
RrC10414_p1 110 RIIVY-NSRSVGLKKTLVFYRGRAPNGERTDWVMHEYTM 147
****9.999****************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.11E-53 | 12 | 170 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 51.879 | 12 | 170 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.5E-25 | 14 | 146 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900057 | Biological Process | positive regulation of leaf senescence | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 581 aa Download sequence Send to blast |
MANSCLKGGR FSAPGFRFHP TDEELVVYYL KRKICGRKLR INVIGVVDVY KVDPTELPKL 60 NNLCEAGLSV LKSGDRQWFY FTPRNRKYPN AARSSRGTAT GYWKATGKDR IIVYNSRSVG 120 LKKTLVFYRG RAPNGERTDW VMHEYTMDEE ELGRCKNAKE YYALYKLYKK SGAGPKNGEE 180 YGAPFQEEEW VDGDDDDGEE ADDVAVPDDH VVRYENNPRV DNGLFCNPVN VRADDFNKLL 240 NEIPYAPGLA PRQMNEVSGV MQVNSQEEMQ STLLNNSTGE FLAPWEVGMF LPNCQPNSIH 300 SSYQSHVRSA DSFDCSEVKS AFKTSGVAPF VFEKEDYIEM DDLLTPEQVA VSVEKPGQFL 360 NPGEEYEHFN DYDQLFHDVS MSFDLEPVFQ ETYADLSSVS NFANDGRQPF LYQQQFQDQT 420 LETQANSFMD PNPNANQLVD DVWFKDAQAA LYDQPQSSSL AFASPSSGLM PESKNQECQN 480 GGETRSQFSS AMWALLDSVP STPASACEGP LNRTFVRMSS LSRIRFNGTS VTTSKVTVAK 540 KSFGNKGFLL LSVMGAMCAF FWVFIIAIVG VQGRHRYLNM S |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 4e-43 | 14 | 147 | 17 | 140 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP) (By similarity). Transcriptional activator that promotes leaf senescence by up-regulating senescence-associated genes in response to developmental and stress-induced senescence signals. Functions in salt and oxidative stress-responsive signaling pathways. Binds to the promoter of NAC029/NAP and NAC059/ORS1 genes (PubMed:23926065). {ECO:0000250|UniProtKB:Q949N0, ECO:0000269|PubMed:23926065}. | |||||
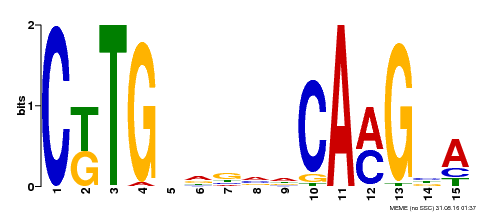
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00178 | DAP | Transfer from AT1G34180 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold, salt, drought stress and methyl methanesulfonate (MMS) treatment. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018487250.1 | 0.0 | PREDICTED: NAC domain-containing protein 16-like | ||||
| Swissprot | A4FVP6 | 0.0 | NAC16_ARATH; NAC domain-containing protein 16 | ||||
| TrEMBL | A0A0D3CGH3 | 0.0 | A0A0D3CGH3_BRAOL; Uncharacterized protein | ||||
| STRING | Bo5g085440.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2812 | 26 | 67 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G34180.1 | 0.0 | NAC domain containing protein 16 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



