 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC12529_p1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 282aa MW: 32541.3 Da PI: 5.2593 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 159.4 | 1.4e-49 | 6 | 139 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk. 95
lppGfrFhPtdeelv +yL+++ eg ++el e+i+ +d+yk++Pw+Lp k + +++ ew+fF++rd+ky++g+r nratksgyWkatgkd++++++
RrC12529_p1 6 LPPGFRFHPTDEELVGYYLHRRNEGLEIEL-EIIPLMDLYKFDPWELPeKsFLPNRDMEWFFFCHRDRKYQNGSRINRATKSGYWKATGKDRNIICQs 102
79****************************.99**************95334556888**************************************85 PP
NAM 96 .kgel.....vglkktLvfykgrapkgektdWvmheyrl 128
+ g++ktLvfy grap g +t+W mheyrl
RrC12529_p1 103 sS--SssspiSGCRKTLVFYLGRAPFGGRTEWAMHEYRL 139
51..3455669**************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.84E-56 | 3 | 163 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 55.318 | 6 | 163 | IPR003441 | NAC domain |
| Pfam | PF02365 | 5.7E-27 | 7 | 139 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 282 aa Download sequence Send to blast |
MARSYLPPGF RFHPTDEELV GYYLHRRNEG LEIELEIIPL MDLYKFDPWE LPEKSFLPNR 60 DMEWFFFCHR DRKYQNGSRI NRATKSGYWK ATGKDRNIIC QSSSSSSSPI SGCRKTLVFY 120 LGRAPFGGRT EWAMHEYRLF DSDSSEKSRN FKGDFALCRV VKRNEVTLKK CEASSLEVPD 180 DPLANNVDIP CEARYTGTGP CDAHNTLRAS PDFILESSTK GNSQSQTEEN SGFQVFALPE 240 FDYPSEFFAD LNFDRGMENP FTFVNYQEAH VNDEVINYGV LR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 8e-48 | 6 | 169 | 17 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 8e-48 | 6 | 169 | 17 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 8e-48 | 6 | 169 | 17 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 8e-48 | 6 | 169 | 17 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 8e-48 | 6 | 169 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 8e-48 | 6 | 169 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 8e-48 | 6 | 169 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 8e-48 | 6 | 169 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 8e-48 | 6 | 169 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 8e-48 | 6 | 169 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 8e-48 | 6 | 169 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 8e-48 | 6 | 169 | 20 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 8e-48 | 6 | 169 | 17 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 8e-48 | 6 | 169 | 17 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in tissue reunion of wounded inflorescence stems. Required for the division of pith cells in the reunion process, which is dependent on polar-transported auxin and the wound-inducible hormones ethylene and jasmonate (PubMed:21911380). Binds to the promoters of XTH19 and XTH20 to induce their expression via auxin signaling. XTH19 and XTH20 are involved in cell proliferation in the tissue reunion process of incised stems (PubMed:25182467). Involved in hypocotyl graft union formation. Required for the auxin- mediated promotion of vascular tissue proliferation during hypocotyl graft attachment (PubMed:27986917). {ECO:0000269|PubMed:21911380, ECO:0000269|PubMed:25182467, ECO:0000269|PubMed:27986917}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00175 | DAP | Transfer from AT1G32510 | Download |
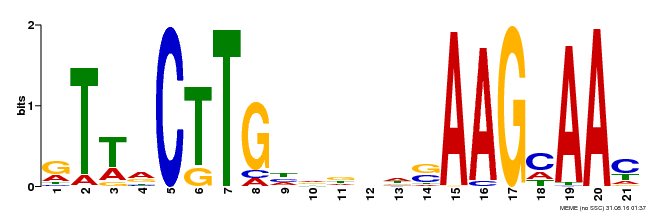
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding in the flowering stem. {ECO:0000269|PubMed:21911380}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP002684 | 2e-78 | CP002684.1 Arabidopsis thaliana chromosome 1 sequence. | |||
| GenBank | F5D14 | 2e-78 | AC007767.3 Sequence of BAC F5D14 from Arabidopsis thaliana chromosome 1, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018449878.1 | 0.0 | PREDICTED: NAC domain-containing protein 86 | ||||
| Swissprot | O49697 | 2e-86 | NAC71_ARATH; NAC domain-containing protein 71 | ||||
| TrEMBL | A0A0D3DKT5 | 0.0 | A0A0D3DKT5_BRAOL; Uncharacterized protein | ||||
| STRING | Bo8g028730.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM15029 | 17 | 19 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32510.1 | 1e-147 | NAC domain containing protein 11 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



