 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC12761_p2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 280aa MW: 31889.1 Da PI: 8.5722 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 90 | 1.2e-28 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krien rqvtfskRr+g+lKKA+ELSvLCd evavi+fs++gkl+e+ss
RrC12761_p2 9 KRIENANSRQVTFSKRRAGLLKKAHELSVLCDSEVAVIVFSKSGKLFEFSS 59
79***********************************************96 PP
| |||||||
| 2 | K-box | 74.5 | 3.1e-25 | 88 | 176 | 12 | 100 |
K-box 12 akaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkklee 100
+ +e+ ++e++ Lk+ei++Lq+++ +++G++L+ L lkeLq+LeqqL+ sl ++R++K+ ll++q+ee++ ke++++ en++Lr++++e
RrC12761_p2 88 NLQEEDCTEVDLLKSEISKLQEKHLQMQGKGLNALCLKELQHLEQQLNLSLISVRERKELLLTKQLEESRLKEQRAELENETLRRQVQE 176
5578899*******************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 32.59 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 2.8E-42 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 4.19E-31 | 2 | 76 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.04E-41 | 2 | 74 | No hit | No description |
| PRINTS | PR00404 | 1.5E-29 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 3.9E-26 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.5E-29 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.5E-29 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.3E-16 | 89 | 174 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 13.105 | 90 | 180 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010262 | Biological Process | somatic embryogenesis | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048577 | Biological Process | negative regulation of short-day photoperiodism, flowering | ||||
| GO:0060862 | Biological Process | negative regulation of floral organ abscission | ||||
| GO:0060867 | Biological Process | fruit abscission | ||||
| GO:0071365 | Biological Process | cellular response to auxin stimulus | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 280 aa Download sequence Send to blast |
MGRGKIEIKR IENANSRQVT FSKRRAGLLK KAHELSVLCD SEVAVIVFSK SGKLFEFSST 60 KCMKKTLLRY SNYNPSPDAP LINYKPENLQ EEDCTEVDLL KSEISKLQEK HLQMQGKGLN 120 ALCLKELQHL EQQLNLSLIS VRERKELLLT KQLEESRLKE QRAELENETL RRQVQELRSF 180 LPSINRNYVP SYITCFAIDP KNSLVNNSGL DDTNYSLQKT NSDTTLQLGL PGEAQARRRS 240 EGNRESPSSD SATTSTTNCT EDHLKTKANG SPFSHREIWE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 2e-23 | 1 | 74 | 1 | 73 | MEF2C |
| 5f28_B | 2e-23 | 1 | 74 | 1 | 73 | MEF2C |
| 5f28_C | 2e-23 | 1 | 74 | 1 | 73 | MEF2C |
| 5f28_D | 2e-23 | 1 | 74 | 1 | 73 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00508 | DAP | Transfer from AT5G13790 | Download |
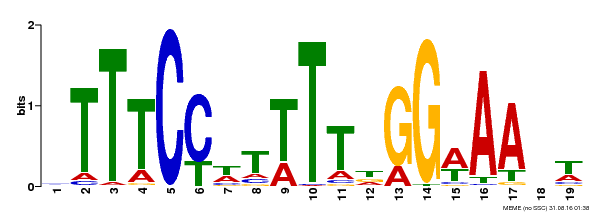
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BNU22681 | 0.0 | U22681.1 Brassica napus AGL15 type 2 mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018444335.1 | 0.0 | PREDICTED: agamous-like MADS-box protein AGL15 | ||||
| Swissprot | Q39295 | 1e-138 | AGL15_BRANA; Agamous-like MADS-box protein AGL15 | ||||
| TrEMBL | A0A0D3EFP6 | 1e-158 | A0A0D3EFP6_BRAOL; Uncharacterized protein | ||||
| STRING | Bo9g166450.1 | 1e-159 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7518 | 25 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13790.1 | 1e-120 | AGAMOUS-like 15 | ||||



