 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC1613_p1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 407aa MW: 45826.5 Da PI: 7.7252 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.2 | 1.4e-31 | 80 | 133 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtpeLH rF++ave+LGG+++AtPk +l+lm+vkgL ++hvkSHLQ+YR+
RrC1613_p1 80 PRLRWTPELHLRFLQAVERLGGPDRATPKLVLQLMNVKGLGIAHVKSHLQMYRS 133
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 6.6E-30 | 76 | 134 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 12.139 | 76 | 136 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.87E-15 | 77 | 135 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-23 | 80 | 135 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 8.3E-9 | 81 | 132 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 407 aa Download sequence Send to blast |
MRSSSQSSEN SKTCPSNKFK TTTKKEDNRD DEDEDEEERS VDQSPSRNSY VEESGSHHHN 60 NDQIKKNGGS VRPYNRSKTP RLRWTPELHL RFLQAVERLG GPDRATPKLV LQLMNVKGLG 120 IAHVKSHLQM YRSKKTDDPN QGDQGFSFEH GGGYTCNLGQ LPMLQSFDQR PSTSLGYGGG 180 SWIDHRRQVY RSPWRGLTAR DSTRTRQTLF SSQSGERFRG ISNNILDDKN KTILFRINSH 240 EAAQENNGVA EAVPSIHRSF LEGMKTLSKS WGQSPSSIPN SSTASRPQDL CATTLSSNQK 300 ENLRVAEETE NVLKRKRLSL SDDCNKSDQD LDLSLSLKVP RTHNNLGDFL LEDEEKEHDD 360 HEDIKGLSLS LSSSSSSKFD QVIRKEDQND PKTRNISVLA SPLDLTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 3e-17 | 81 | 135 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 3e-17 | 81 | 135 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 3e-17 | 81 | 135 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 3e-17 | 81 | 135 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
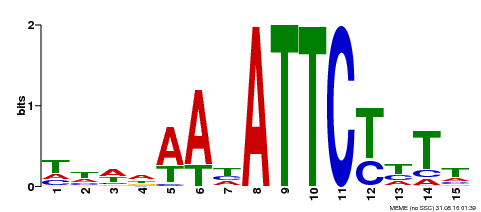
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018458721.1 | 0.0 | PREDICTED: uncharacterized protein LOC108829586 | ||||
| TrEMBL | A0A0D3BQA8 | 0.0 | A0A0D3BQA8_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3P6BI71 | 0.0 | A0A3P6BI71_BRAOL; Uncharacterized protein | ||||
| STRING | Bo4g025360.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9359 | 21 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40260.1 | 1e-178 | G2-like family protein | ||||



