 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC17018_p1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 482aa MW: 54408.9 Da PI: 6.7725 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 38.9 | 1.6e-12 | 7 | 66 | 23 | 96 |
.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEE CS
B3 23 aeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkv 96
+++h + l + d s + W+vkl ++++ l++GW+eF+ka+ ++gD +vF++dg++ f +v++
RrC17018_p1 7 LRNHTK------VLLISDASDKIWKVKL--EGNR----LAGGWEEFAKAHSFRDGDLLVFRHDGDEIF--HVSA 66
544443......477889**********..9998....************************998888..7665 PP
| |||||||
| 2 | B3 | 65.3 | 8.8e-21 | 127 | 224 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE- CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfr 98
f+ + +++++l ++rl l+k+f+ ++++++ ++ l + +gr+W++ l+ + +sg +++++GW +F++angL +gD++ Fkl + +e+++v+++++
RrC17018_p1 127 FLRARVTPYSLIKDRLDLSKDFTVVSFNEHNKPCEIDLVNDKGRKWTLLLSKNTSSGVFYIRRGWVNFCSANGLSQGDICKFKLAE-DEERPVLRLCS 223
667788999*************7666444556669***************99999999*************************987.66669999998 PP
S CS
B3 99 k 99
+
RrC17018_p1 224 S 224
6 PP
| |||||||
| 3 | B3 | 65.5 | 7.9e-21 | 271 | 368 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTE..EEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgr..yvltkGWkeFvkangLkegDfvvFkldgrsefelvvkv 96
f+++ ++++ l++g+l+++ f++e g+kk s ++tl +++gr+W +l+++++ g+ ++l+ GW+e++kang+k++D++v++l+ +++++v+k+
RrC17018_p1 271 FLTIKLTPNRLQTGQLYISSVFINESGIKK--SGEITLLNQDGRKWPSYLQMTGQCGSewFYLRHGWREMCKANGVKVNDSFVLELIC-EDANPVLKF 365
6778889999*************9999885..459***************9966655556***************************9.9999***** PP
E-S CS
B3 97 frk 99
++k
RrC17018_p1 366 CSK 368
987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50863 | 12.323 | 1 | 70 | IPR003340 | B3 DNA binding domain |
| CDD | cd10017 | 1.02E-9 | 6 | 65 | No hit | No description |
| SMART | SM01019 | 4.9E-6 | 6 | 70 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 9.42E-12 | 6 | 66 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 1.7E-7 | 10 | 64 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.3E-11 | 11 | 66 | IPR015300 | DNA-binding pseudobarrel domain |
| Pfam | PF02362 | 7.4E-19 | 127 | 224 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 9.4E-25 | 127 | 225 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.734 | 127 | 225 | IPR003340 | B3 DNA binding domain |
| SuperFamily | SSF101936 | 2.35E-17 | 129 | 225 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 7.7E-17 | 131 | 224 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.02E-17 | 137 | 222 | No hit | No description |
| Gene3D | G3DSA:2.40.330.10 | 3.8E-18 | 267 | 368 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 5.1E-18 | 269 | 368 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.18E-14 | 270 | 367 | No hit | No description |
| PROSITE profile | PS50863 | 12.816 | 271 | 369 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 2.4E-21 | 271 | 368 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 9.0E-25 | 271 | 369 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 482 aa Download sequence Send to blast |
RLGDEFLRNH TKVLLISDAS DKIWKVKLEG NRLAGGWEEF AKAHSFRDGD LLVFRHDGDE 60 IFHVSASPRS NSCDTSHHAS PSFVDTDDAE SDDEYSESDE EEEECDDDAG DILVNKKKKT 120 EGGFSCFLRA RVTPYSLIKD RLDLSKDFTV VSFNEHNKPC EIDLVNDKGR KWTLLLSKNT 180 SSGVFYIRRG WVNFCSANGL SQGDICKFKL AEDEERPVLR LCSSSNSHEE EEECLEADAA 240 KIGSVGGCSK EKRRLDECSK RKKKNNNPSK FLTIKLTPNR LQTGQLYISS VFINESGIKK 300 SGEITLLNQD GRKWPSYLQM TGQCGSEWFY LRHGWREMCK ANGVKVNDSF VLELICEDAN 360 PVLKFCSKVD NKGNGNRRAC KRRAHEEAPI VETEGRKRGR PRLSNRHATN SSHLRRTQQE 420 SCSVADQVAN VKLSILDTLN TVRQFRAELE AREKNLEATL LEVDSLGDRI LGISQSLTSN 480 NV |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 249 | 262 | KEKRRLDECSKRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May play a role in flower development. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00602 | PBM | Transfer from AT4G31610 | Download |
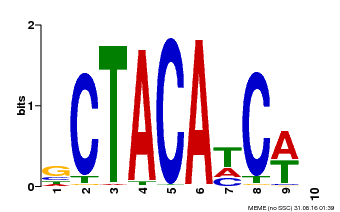
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018447207.1 | 0.0 | PREDICTED: B3 domain-containing protein REM1-like | ||||
| Swissprot | O65098 | 1e-180 | REM1_BRAOB; B3 domain-containing protein REM1 | ||||
| TrEMBL | M4D179 | 0.0 | M4D179_BRARP; Uncharacterized protein | ||||
| STRING | Bra010228.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9950 | 17 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31610.1 | 1e-154 | B3 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



