 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC2041_p2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 343aa MW: 38925.1 Da PI: 7.3635 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 99.5 | 2.3e-31 | 206 | 260 | 2 | 56 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
+r+rW++eLH++Fv+a+++LGG+++AtPk+i+e+m v+gLt+++vkSHLQkYR++
RrC2041_p2 206 QRRRWSQELHRKFVDALHSLGGPQVATPKQIREMMRVDGLTNDEVKSHLQKYRMH 260
8****************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-27 | 203 | 263 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 4.17E-17 | 203 | 263 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.054 | 203 | 262 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 1.3E-24 | 206 | 260 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 5.2E-8 | 207 | 258 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010092 | Biological Process | specification of organ identity | ||||
| GO:0010629 | Biological Process | negative regulation of gene expression | ||||
| GO:0048449 | Biological Process | floral organ formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 343 aa Download sequence Send to blast |
MVQTDTDKMG LNLNLSMYSL AKPLSQFLDQ VSRIKDHDSK LSEIDDYVGK LEEERRKIDV 60 FKRELPLCML LLNEAIERLK EEASSVMMMA SNGKLDVGEG VILESDNNKK NWMSSAQLWI 120 SNPNSQLQST NEEEDGCVTQ KPIQTCNNQG GAFPPPYNPP PPPPPAPLSL RTPTSEILMD 180 YRRIEQNHPQ FSKPIIQSHH VPKKDQRRRW SQELHRKFVD ALHSLGGPQV ATPKQIREMM 240 RVDGLTNDEV KSHLQKYRMH IRKHPLHPAK TLPSSDQAVL LDRETQSLIS LTRSDSPQSP 300 LVVDRGCLFS NNGHSSEDEV KSDGRSSWKS VSNKNRPALD LEL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 6e-14 | 207 | 258 | 4 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 6e-14 | 207 | 258 | 4 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 6e-14 | 207 | 258 | 3 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 6e-14 | 207 | 258 | 3 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 6e-14 | 207 | 258 | 3 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 6e-14 | 207 | 258 | 3 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 6e-14 | 207 | 258 | 4 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 6e-14 | 207 | 258 | 4 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 6e-14 | 207 | 258 | 4 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 6e-14 | 207 | 258 | 4 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 6e-14 | 207 | 258 | 4 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 6e-14 | 207 | 258 | 4 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 201 | 207 | PKKDQRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
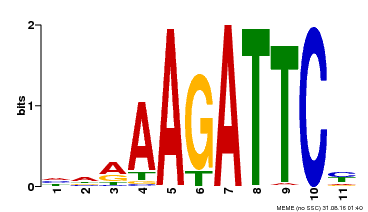
| UniProt | Transcriptional repressor that functions with ULT1 in a pathway which regulates floral meristem homeostasis and organ number in the flower. Binds specifically to the DNA sequence motif 5'-GTAGATTCCT-3' of WUS promoter, and may be involved in direct regulation of WUS expression. Binds specifically to the DNA sequence motif 5'-AAGAATCTTT-3' found in the promoters of AG and the NAC domain genes CUC1, CUC2 and CUC3, and may be involved in direct regulation of these gene expressions. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00475 | DAP | Transfer from AT4G37180 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018453470.1 | 0.0 | PREDICTED: myb family transcription factor EFM | ||||
| Refseq | XP_018453831.1 | 0.0 | PREDICTED: myb family transcription factor EFM-like | ||||
| Swissprot | F4JRB0 | 1e-159 | HHO5_ARATH; Transcription factor HHO5 | ||||
| TrEMBL | A0A398AHG2 | 0.0 | A0A398AHG2_BRACM; Uncharacterized protein | ||||
| STRING | Bo1g005280.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4922 | 26 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37180.1 | 1e-164 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



