 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC3673_p2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 276aa MW: 30670.2 Da PI: 7.2321 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 96.9 | 1.5e-30 | 82 | 135 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
+prl+Wtp+LH+rFv+av++L G+++A+Pkti++lm+v+gLt+e+v+SHLQkYRl
RrC3673_p2 82 RPRLVWTPQLHKRFVDAVAHL-GIKNAVPKTIMQLMSVDGLTRENVASHLQKYRL 135
59*******************.********************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.84E-20 | 79 | 139 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.059 | 79 | 138 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-30 | 81 | 140 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.8E-24 | 83 | 135 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.0E-7 | 85 | 134 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 276 aa Download sequence Send to blast |
MREEDSNWFA KWEEELPSPE ELIPLSQSLI TPDLAIAFDL RTPNQNSGQP QPHQTTPPQQ 60 TNSSAEFAAD STGDEPARTL KRPRLVWTPQ LHKRFVDAVA HLGIKNAVPK TIMQLMSVDG 120 LTRENVASHL QKYRLYLKRM QSGGGGGGGS VGDSDHLFAS SPVPTHFLHP TARQSSDHFI 180 PSFVPIAALQ HQHRQQQPPP QFLHRQISAV NFTSQGNNSG KAMDQSLFLA RQSQQQQQQV 240 TARPSSLHLH SQVANYADDL EQGANTVLTL FPTREN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5lxu_A | 1e-31 | 82 | 138 | 1 | 57 | Transcription factor LUX |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that acts as a negative regulator of freezing tolerance via a CBF-independent pathway. {ECO:0000269|PubMed:20331973}. | |||||
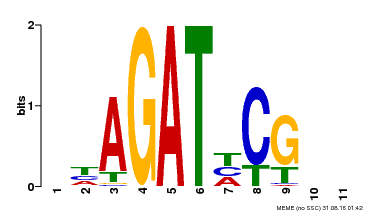
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00056 | PBM | Transfer from AT5G05090 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by drought, salt and abscisic acid (ABA). Down-regulated by cold. {ECO:0000269|PubMed:20331973}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018442772.1 | 0.0 | PREDICTED: transcription factor PCL1-like | ||||
| Swissprot | O22210 | 7e-60 | MYBC1_ARATH; Transcription factor MYBC1 | ||||
| TrEMBL | A0A397XQK4 | 1e-142 | A0A397XQK4_BRACM; Uncharacterized protein | ||||
| TrEMBL | M4CXZ0 | 1e-142 | M4CXZ0_BRARP; Uncharacterized protein | ||||
| STRING | Bra009087.1-P | 1e-143 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1549 | 27 | 91 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G05090.1 | 1e-108 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



