 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC5036_p1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 375aa MW: 40473.1 Da PI: 7.7959 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 70.5 | 2.6e-22 | 268 | 330 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkr++rkq+NRe+ArrsR+RK+ae++eL++++++L +eN +L+ e+++lk ++++l +e+
RrC5036_p1 268 ERELKRQKRKQSNRESARRSRLRKQAECDELAQRADVLNGENASLRAEINKLKSQYEELLAEN 330
89*********************************************************9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 1.7E-27 | 22 | 104 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 1.4E-11 | 139 | 175 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 1.1E-17 | 260 | 326 | No hit | No description |
| SMART | SM00338 | 1.0E-20 | 268 | 332 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 9.4E-21 | 268 | 330 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 12.668 | 270 | 333 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 6.13E-11 | 271 | 327 | No hit | No description |
| CDD | cd14702 | 7.66E-24 | 273 | 323 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 275 | 290 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 375 aa Download sequence Send to blast |
MVGSEMEKTG QENEPKTPPP PAPPVTSQEP SSSSAVSAGM STTPDWSGFQ AYSPMPPHGY 60 VASSPQPHPY MWGVQHMMPP YGTPPHPYVA MYPPGGMYGH PSLPPGSYPY SPYAMPSPNG 120 MAEASGNTEG DGKQAEVKEK LPIKRSKGSL GSLKMIIGKN NETGKNSGAS ANGALLRALL 180 MVQVKEVMQI HKTNNGKDGS DSAHGPPPRN GSNQPVNQIV PIMPTGVPGP PTNLNIGMEY 240 WSGHGNVSTT VPGVVVDGSQ SQTWIQDERE LKRQKRKQSN RESARRSRLR KQAECDELAQ 300 RADVLNGENA SLRAEINKLK SQYEELLAEN NSLKNRFTAV PSQEGVNLDK KEQEPEASTR 360 QDVAETTYGS YNNSA |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 284 | 290 | RRSRLRK |
| 2 | 284 | 291 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds to the G-box motif (5'-CACGTG-3') and other cis-acting elements with 5'-ACGT-3' core, such as Hex, C-box and as-1 motifs. Possesses high binding affinity to G-box, much lower affinity to Hex and C-box, and little affinity to as-1 element (PubMed:18315949). G-box and G-box-like motifs are cis-acting elements defined in promoters of certain plant genes which are regulated by such diverse stimuli as light-induction or hormone control (Probable). Binds to the G-box motif 5'-CACGTG-3' of LHCB2.4 (At3g27690) promoter. May act as transcriptional activator in light-regulated expression of LHCB2.4. Probably binds DNA as monomer. DNA-binding activity is redox-dependent (PubMed:22718771). {ECO:0000269|PubMed:18315949, ECO:0000269|PubMed:22718771, ECO:0000305|PubMed:18315949}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00173 | DAP | Transfer from AT1G32150 | Download |
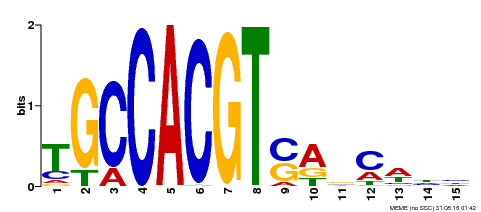
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353505 | 1e-153 | AK353505.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-47-I14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018447676.1 | 0.0 | PREDICTED: bZIP transcription factor 68 | ||||
| Swissprot | Q84LG2 | 0.0 | BZP68_ARATH; bZIP transcription factor 68 | ||||
| TrEMBL | A0A078FG90 | 0.0 | A0A078FG90_BRANA; BnaA09g24610D protein | ||||
| TrEMBL | A0A397XX62 | 0.0 | A0A397XX62_BRACM; Uncharacterized protein | ||||
| STRING | Bo5g048660.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3701 | 25 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32150.1 | 1e-167 | basic region/leucine zipper transcription factor 68 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



