 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC5155_p4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 510aa MW: 56687.8 Da PI: 5.2871 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 152 | 2.7e-47 | 15 | 140 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkg 97
l+pGfrFhPtdeelvv+yLk+k+++kkl++ +i e+d+yk++P++Lp k +++++++w+fFs rd+k+ g r++rat +gyWkatg d+ +++ ++
RrC5155_p4 15 LAPGFRFHPTDEELVVYYLKRKTRRKKLRV-AAIGETDVYKFDPEELPgKaLYNTGDRQWFFFSLRDRKQ--GGRSSRATGHGYWKATGVDRVIKC-NS 109
579***************************.88***************7435667999**********86..89**********************.99 PP
NAM 98 elvglkktLvfykgrapkgektdWvmheyrl 128
+ vg kktLvf++grapkge+t+Wvmhey+l
RrC5155_p4 110 RPVGEKKTLVFHRGRAPKGERTNWVMHEYTL 140
9****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.7E-54 | 11 | 163 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 51.717 | 15 | 163 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.6E-25 | 17 | 140 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010114 | Biological Process | response to red light | ||||
| GO:0010224 | Biological Process | response to UV-B | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:1900409 | Biological Process | positive regulation of cellular response to oxidative stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 510 aa Download sequence Send to blast |
METSRGSRPA GDRVLAPGFR FHPTDEELVV YYLKRKTRRK KLRVAAIGET DVYKFDPEEL 60 PGKALYNTGD RQWFFFSLRD RKQGGRSSRA TGHGYWKATG VDRVIKCNSR PVGEKKTLVF 120 HRGRAPKGER TNWVMHEYTL HKEELKMCGD VKHNYVLYKI FKKSGSGPKN GEQYGAPFVE 180 EEWAEDDDEV DEADAVVVPA NQLMVSAGMG NNNNIWADGG GLNQSELNEN DIQELMRQVS 240 EETGVNSHVA NDNPVNLSED EYLEIDDLLL PGPEPSYVDQ EESAVLNDNN FFDVDSYIGD 300 FDATNPQTEP VGVVGLNNGV VHSLPVADQA NNNQFQQQTW KNQDSNWPLR NSYTRKINSG 360 SWTHELNNDE VTVCGYGEAP GTGDASEFTN PLTSALSTAK GEEATKDQSS QFTSSVWSFL 420 ESMPASPAFA SESPIVNLNI VRISSLGGRY RFNSKSTSSN VVVRKKSGEN NTNKKKNHKG 480 FLCLSIIGAL CALSWVMLGT MGVSGRSLLW |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 5e-44 | 9 | 169 | 11 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 5e-44 | 9 | 169 | 11 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 5e-44 | 9 | 169 | 11 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 5e-44 | 9 | 169 | 11 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 5e-44 | 9 | 169 | 14 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 5e-44 | 9 | 169 | 14 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 5e-44 | 9 | 169 | 14 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 5e-44 | 9 | 169 | 14 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 5e-44 | 9 | 169 | 14 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 5e-44 | 9 | 169 | 14 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 5e-44 | 9 | 169 | 14 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 5e-44 | 9 | 169 | 14 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 5e-44 | 9 | 169 | 11 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 5e-44 | 9 | 169 | 11 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
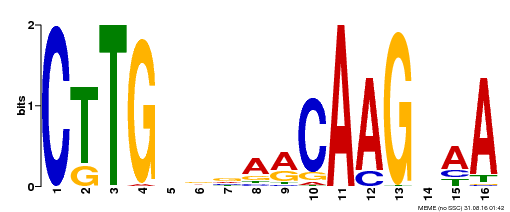
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP). Involved in oxidative stress tolerance by mediating regulation of mitochondrial retrograde signaling during mitochondrial dysfunction. Interacts directly with the mitochondrial dysfunction DNA consensus motif 5'-CTTGNNNNNCA[AC]G-3', a cis-regulatory elements of several mitochondrial retrograde regulation-induced genes, and triggers increased oxidative stress tolerance. {ECO:0000269|PubMed:24045019}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00176 | DAP | Transfer from AT1G32870 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat, salt stress, abscisic acid (ABA) and methyl methanesulfonate (MMS) treatment. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF974756 | 0.0 | KF974756.1 Brassica napus NAC transcription factor 13-1 (NAC13-1.1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018441206.1 | 0.0 | PREDICTED: NAC domain-containing protein 13 | ||||
| Swissprot | F4IED2 | 0.0 | NAC13_ARATH; NAC domain-containing protein 13 | ||||
| TrEMBL | A0A397YRI4 | 0.0 | A0A397YRI4_BRACM; Uncharacterized protein | ||||
| STRING | Bo7g057730.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM15317 | 15 | 17 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32870.1 | 0.0 | NAC domain protein 13 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



