 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC5322_p2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 290aa MW: 33321.6 Da PI: 6.0823 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 64.9 | 1.3e-20 | 109 | 168 | 3 | 59 |
-SS-EEEEEEE--TT-SS-EEEEEE-ST...T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 3 DgynWrKYGqKevkgsefprsYYrCtsa...gCpvkkkversaedpkvveitYegeHnhe 59
Dg+ WrKYGqK++k++++ rsYYrC+++ C ++k+ ++ +++p v +tY g+H+++
RrC5322_p2 109 DGFLWRKYGQKSIKNNKYERSYYRCSYNidhACGARKHEQQIKDNPPVYRTTYFGHHTCK 168
9**************************99999**************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 19.187 | 102 | 170 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.5E-22 | 102 | 168 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.75E-21 | 104 | 169 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.8E-30 | 107 | 169 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.5E-21 | 109 | 168 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009863 | Biological Process | salicylic acid mediated signaling pathway | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 290 aa Download sequence Send to blast |
MNSPHEKAVQ AILYGHSCAK RLKRRLEDPM ADDRSISNYD LATSIVHCFS SAISILSDKP 60 ISEDDQVSDL SSMDSSPPLL QRNHSKRRKI NSTNSTKNWR DDSPDPYYDG FLWRKYGQKS 120 IKNNKYERSY YRCSYNIDHA CGARKHEQQI KDNPPVYRTT YFGHHTCKIN NNHDSVFTAV 180 RDQVDVVGSS RIIQFGKELD QEKGSCLTDF SLSVKHEESI IKEETTDHYR EITGDDKDCQ 240 HVMDENQSSL SSSYTPPSSS VSETDMFDTD LLLNNLDSWD RYGLFDFGVQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 2e-15 | 109 | 168 | 10 | 69 | OsWRKY45 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
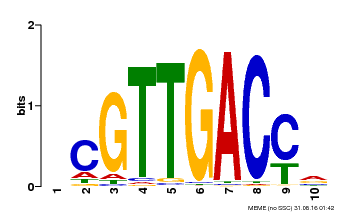
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00619 | PBM | Transfer from AT5G22570 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018444308.1 | 0.0 | PREDICTED: probable WRKY transcription factor 38 | ||||
| Swissprot | Q8GWF1 | 1e-120 | WRK38_ARATH; Probable WRKY transcription factor 38 | ||||
| TrEMBL | A0A3P6CS12 | 0.0 | A0A3P6CS12_BRACM; Uncharacterized protein | ||||
| STRING | Bra002421.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13673 | 17 | 27 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G22570.1 | 1e-117 | WRKY DNA-binding protein 38 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



