 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC5791_p3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 226aa MW: 25976 Da PI: 7.6922 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 125.8 | 3.4e-39 | 5 | 132 | 3 | 128 |
NAM 3 pGfrFhPtdeelvveyLkkkvegkk..leleevikevdiykvePwdLp..kkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk.. 95
+G+rF Ptdee+v++yL+ k +g + ++++vi++++i++++PwdLp ++++++++ wyfF +++ ky++g++++r+tksg+Wk+tg + +++k
RrC5791_p3 5 VGLRFLPTDEEIVDHYLRLKNNGGSntSHVDQVISTLNICNFDPWDLPpkSRIESSNQVWYFFGRKEGKYKRGEQQKRQTKSGFWKKTGVTLLIKRKrg 103
7*******************9998866677789**************86448888999************************************99977 PP
NAM 96 kgelvglkktLvfykgrapkgektdWvmheyrl 128
++e++g k++Lvfy ++g k++Wvmhey+
RrC5791_p3 104 NREKIGEKRVLVFY----KNGYKSNWVMHEYHH 132
7888**********....7899*********95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 40.428 | 3 | 152 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 1.96E-40 | 4 | 152 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.3E-23 | 5 | 131 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 226 aa Download sequence Send to blast |
MENMVGLRFL PTDEEIVDHY LRLKNNGGSN TSHVDQVIST LNICNFDPWD LPPKSRIESS 60 NQVWYFFGRK EGKYKRGEQQ KRQTKSGFWK KTGVTLLIKR KRGNREKIGE KRVLVFYKNG 120 YKSNWVMHEY HHTSFLYPNE SGKYTICKVQ FKGEVSEISS SSGVDNVENS HSLATPTNNP 180 GGSEGLQNSS QFSGFLDHQQ EMQFEAETQI DFNDLLSYDL ESLLAS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 2e-21 | 6 | 149 | 20 | 162 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 2e-21 | 6 | 149 | 20 | 162 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 2e-21 | 6 | 149 | 20 | 162 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 2e-21 | 6 | 149 | 20 | 162 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 2e-21 | 6 | 149 | 23 | 165 | NAC domain-containing protein 19 |
| 3swm_B | 2e-21 | 6 | 149 | 23 | 165 | NAC domain-containing protein 19 |
| 3swm_C | 2e-21 | 6 | 149 | 23 | 165 | NAC domain-containing protein 19 |
| 3swm_D | 2e-21 | 6 | 149 | 23 | 165 | NAC domain-containing protein 19 |
| 3swp_A | 2e-21 | 6 | 149 | 23 | 165 | NAC domain-containing protein 19 |
| 3swp_B | 2e-21 | 6 | 149 | 23 | 165 | NAC domain-containing protein 19 |
| 3swp_C | 2e-21 | 6 | 149 | 23 | 165 | NAC domain-containing protein 19 |
| 3swp_D | 2e-21 | 6 | 149 | 23 | 165 | NAC domain-containing protein 19 |
| 4dul_A | 2e-21 | 6 | 149 | 20 | 162 | NAC domain-containing protein 19 |
| 4dul_B | 2e-21 | 6 | 149 | 20 | 162 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
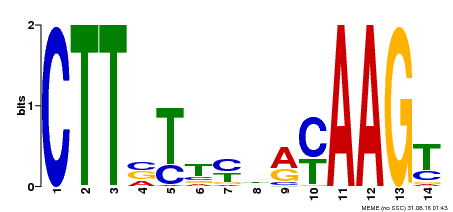
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00122 | DAP | Transfer from AT1G02230 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018490136.1 | 1e-152 | PREDICTED: NAC domain-containing protein 5-like | ||||
| Swissprot | O81913 | 5e-78 | NAC4_ARATH; NAC domain-containing protein 4 | ||||
| Swissprot | O81914 | 7e-78 | NAC5_ARATH; NAC domain-containing protein 5 | ||||
| TrEMBL | A0A078J0S1 | 1e-125 | A0A078J0S1_BRANA; BnaC08g49770D protein | ||||
| STRING | Bo8g117570.1 | 1e-117 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2315 | 16 | 76 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G02250.1 | 6e-75 | NAC domain containing protein 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



