 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC654_p6 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 455aa MW: 50776.2 Da PI: 6.633 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 163.4 | 8.5e-51 | 13 | 141 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp..kkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkge 98
lp+G+rF+Ptdeelv +yL++k++g++ ++ ++i+e+di+k+ePwdLp + +k++++ew fF++ d+ky++g+r+nrat +gyWkatgkd++++s ++
RrC654_p6 13 LPVGLRFRPTDEELVRFYLRRKINGHDDDV-KAIREIDICKWEPWDLPgfSVIKTNDSEWLFFCPLDRKYPNGSRQNRATIAGYWKATGKDRKIKSGRNI 111
799************************999.89***************6448888999**************************************9999 PP
NAM 99 lvglkktLvfykgrapkgektdWvmheyrl 128
+g+k+tLvf++grapkg++t+W+mheyr+
RrC654_p6 112 IIGVKRTLVFHSGRAPKGTRTNWIMHEYRA 141
9***************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.96E-57 | 9 | 164 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 54.867 | 13 | 164 | IPR003441 | NAC domain |
| Pfam | PF02365 | 7.0E-25 | 14 | 140 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009814 | Biological Process | defense response, incompatible interaction | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0070417 | Biological Process | cellular response to cold | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 455 aa Download sequence Send to blast |
MNQNIKVLTL DSLPVGLRFR PTDEELVRFY LRRKINGHDD DVKAIREIDI CKWEPWDLPG 60 FSVIKTNDSE WLFFCPLDRK YPNGSRQNRA TIAGYWKATG KDRKIKSGRN IIIGVKRTLV 120 FHSGRAPKGT RTNWIMHEYR ATEDDLRGTN PGQSPFVICK LFKKQELSLE DEAAEPAVSS 180 PTSSPDETKS EVSVVVKTED VKRYDIAESS FAVSGEATTA KFESQLGDID YLSFPELESL 240 NYTMFSPSHS QLQSELGFSL SAFQSGLSDF SGNYNNSSSS SQVQTQCGTN EVDTYVSEFV 300 DSILDLPDEA VPEQDGSAPH QIAYAQAQHS VGDMSNEVSR TGIIKLQARR EQPSGCATDY 360 IVHGSASKRL RLQSNLDVIK SRSLEIQTIK REVEDTDGDS MKKGKLSKNK TGFLFRKIVS 420 MRCSYGGLFR AAVVAVLFLM SVCSLTADFR ASVMI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 4e-48 | 3 | 165 | 7 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 4e-48 | 3 | 165 | 7 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 4e-48 | 3 | 165 | 7 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 4e-48 | 3 | 165 | 7 | 166 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 3e-48 | 3 | 165 | 10 | 169 | NAC domain-containing protein 19 |
| 3swm_B | 3e-48 | 3 | 165 | 10 | 169 | NAC domain-containing protein 19 |
| 3swm_C | 3e-48 | 3 | 165 | 10 | 169 | NAC domain-containing protein 19 |
| 3swm_D | 3e-48 | 3 | 165 | 10 | 169 | NAC domain-containing protein 19 |
| 3swp_A | 3e-48 | 3 | 165 | 10 | 169 | NAC domain-containing protein 19 |
| 3swp_B | 3e-48 | 3 | 165 | 10 | 169 | NAC domain-containing protein 19 |
| 3swp_C | 3e-48 | 3 | 165 | 10 | 169 | NAC domain-containing protein 19 |
| 3swp_D | 3e-48 | 3 | 165 | 10 | 169 | NAC domain-containing protein 19 |
| 4dul_A | 4e-48 | 3 | 165 | 7 | 166 | NAC domain-containing protein 19 |
| 4dul_B | 4e-48 | 3 | 165 | 7 | 166 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP) (PubMed:20156199, PubMed:19947982). Transcriptional activator involved in response to cold stress. Mediates induction of pathogenesis-related (PR) genes independently of salicylic signaling in response to cold. Binds directly to the PR gene promoters and enhances plant resistance to pathogen infection, incorporating cold signals into pathogen resistance responses (PubMed:19947982). Plays a regulatory role in abscisic acid (ABA)-mediated drought-resistance response (PubMed:22967043). {ECO:0000269|PubMed:19947982, ECO:0000269|PubMed:20156199, ECO:0000269|PubMed:22967043}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00398 | DAP | Transfer from AT3G49530 | Download |
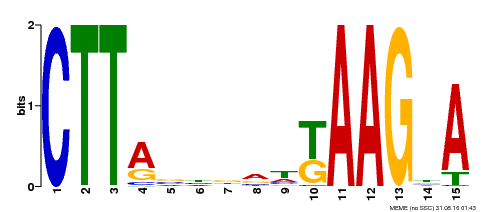
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, drought stress, abscisic acid (ABA), salicylic acid (SA) and methyl methanesulfonate (MMS) treatment. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KP641353 | 0.0 | KP641353.1 Brassica napus NAC transcription factor 62-1.1 (NAC62-1.1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018441133.1 | 0.0 | PREDICTED: NAC domain-containing protein 62-like | ||||
| Swissprot | Q9SCK6 | 0.0 | NAC62_ARATH; NAC domain-containing protein 62 | ||||
| TrEMBL | A0A3P5YGM1 | 0.0 | A0A3P5YGM1_BRACM; Uncharacterized protein | ||||
| TrEMBL | M4DN93 | 0.0 | M4DN93_BRARP; Uncharacterized protein | ||||
| STRING | Bra017980.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2416 | 28 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G49530.1 | 0.0 | NAC domain containing protein 62 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



