 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC67_p2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 427aa MW: 46893 Da PI: 9.8151 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 46.4 | 8.4e-15 | 349 | 400 | 5 | 55 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH.HHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkke.leelkke 55
+r++r++kNRe+A rsR+RK+a++ eLe +v++L+++N++L+ + +e ++k+
RrC67_p2 349 RRQKRMIKNRESAARSRARKQAYTLELEAEVAQLKEQNEELQRKqVEIMEKQ 400
69*************************************9975404434444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 5.4E-14 | 345 | 410 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.921 | 347 | 392 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 6.2E-14 | 349 | 404 | No hit | No description |
| CDD | cd14707 | 1.65E-25 | 349 | 403 | No hit | No description |
| Pfam | PF00170 | 1.1E-12 | 349 | 405 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.5E-10 | 349 | 400 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 352 | 367 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 427 aa Download sequence Send to blast |
MGSRYNFVDE AKQPALGAGV PLMRQNSVFS LTFDEFQNSW GGGIGKDFGS MNMDELLKNI 60 WTAEESHSMM ANNSFNNTVV MNNNGGFSVG VGGELGGGGG GGLQRQGSLT LPRTISQRRV 120 DDVWKELMKE DDTGDGVGNV GGGASGGVPQ RQQTLGEMTL EEFLVRAGVV REESQPVERV 180 DNFNGGFFGF GSDAGLVSAR NGFGGSNQPH GGVVRPDLLT TQTQPLQMQQ QPQKVHQPRE 240 LIPKQDVAFP KQTTIAFSNT VDLVNRSQPP TQEVKPSILG VRDRPMNNNN LLQAVDFKTG 300 VTVAAVSPGS QMSPDLTPKS AMDASLSPVP YMFGRVRKTG AVLEKVIERR QKRMIKNRES 360 AARSRARKQA YTLELEAEVA QLKEQNEELQ RKQVEIMEKQ KKQLLEPMRQ AWGCKRQCLR 420 RTLTGPW |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling. {ECO:0000269|PubMed:11884679, ECO:0000269|PubMed:15361142}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00038 | PBM | Transfer from AT4G34000 | Download |
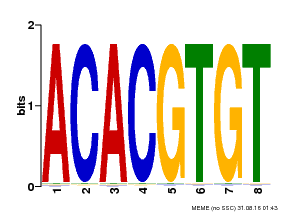
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA). {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:16284313}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353408 | 0.0 | AK353408.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-30-J01. | |||
| GenBank | JQ971973 | 0.0 | JQ971973.1 Eutrema salsugineum abscisic acid responsive elements-binding factor 3 (ABF3) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018450922.1 | 0.0 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 6 | ||||
| Swissprot | Q9M7Q3 | 0.0 | AI5L6_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 6 | ||||
| TrEMBL | E4MY11 | 0.0 | E4MY11_EUTHA; mRNA, clone: RTFL01-30-J01 | ||||
| TrEMBL | V4MBG2 | 0.0 | V4MBG2_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006412286.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM734 | 27 | 129 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G34000.2 | 1e-180 | abscisic acid responsive elements-binding factor 3 | ||||



