 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC7051_p2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 662aa MW: 76152.1 Da PI: 7.4189 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 32.3 | 2.2e-10 | 87 | 128 | 4 | 45 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaL 45
+k rr+++NReAAr+sR+RKka++++Lee +L + ++L
RrC7051_p2 87 DKMKRRLAQNREAARKSRLRKKAYVQQLEESRLKLAQLEQEL 128
7999*************************9655555444443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 2.7E-7 | 78 | 163 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 1.5E-7 | 82 | 131 | No hit | No description |
| PROSITE profile | PS50217 | 9.347 | 86 | 130 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.3E-7 | 87 | 129 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.21E-6 | 88 | 128 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 91 | 106 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 2.8E-28 | 173 | 246 | IPR025422 | Transcription factor TGA like domain |
| SuperFamily | SSF46565 | 1.44E-21 | 455 | 533 | IPR001623 | DnaJ domain |
| Gene3D | G3DSA:1.10.287.110 | 4.0E-21 | 456 | 534 | IPR001623 | DnaJ domain |
| SMART | SM00271 | 1.2E-16 | 463 | 527 | IPR001623 | DnaJ domain |
| PROSITE profile | PS50076 | 17.573 | 464 | 535 | IPR001623 | DnaJ domain |
| CDD | cd06257 | 5.09E-14 | 465 | 524 | IPR001623 | DnaJ domain |
| Pfam | PF00226 | 4.4E-19 | 465 | 532 | IPR001623 | DnaJ domain |
| PRINTS | PR00625 | 5.5E-15 | 466 | 484 | IPR001623 | DnaJ domain |
| PRINTS | PR00625 | 5.5E-15 | 484 | 499 | IPR001623 | DnaJ domain |
| PRINTS | PR00625 | 5.5E-15 | 507 | 527 | IPR001623 | DnaJ domain |
| PROSITE pattern | PS00636 | 0 | 512 | 531 | IPR018253 | DnaJ domain, conserved site |
| PRINTS | PR00625 | 5.5E-15 | 553 | 572 | IPR001623 | DnaJ domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 662 aa Download sequence Send to blast |
MMSSTSPTHL ASLRDMGIYE PFQQLVSWGN VFKSDINDHS PNTASSSVIQ VDHNIKANYP 60 SSSHNQIEVE PSSNDHQEDE DDGMNHDKMK RRLAQNREAA RKSRLRKKAY VQQLEESRLK 120 LAQLEQELEK AKQQGTYSSG SSYVGPSGSI NPSIAAFELE YLHWLEDQSR RVSDIRTALQ 180 ARISDIELRM LVESCLNHYA NLFRMKSDAA KADVFYLISG MWRTSTERLF QWIGGFRPSD 240 LLNVLMPYLQ PLTDQQILEV RNLQQSSQQA EDALSQGIDK LQQSLAENIV IDVVMESSDY 300 PSHMGAAIEN LQALEGFVNQ ADHLRQQTLQ QMAKILTTRQ SARGLLCLGE YLHRLRALSS 360 LWAACPREHA IRSIAEGLKC EKVMSNLRAI CRPHTVFASI VCCSRNQSRS IVKVSIQKLR 420 FRTRVSNSFP SSSRATSKDS NFWFGVSQRK TLVRAASNWS EEKSPYDTLE LERDAEEEQI 480 KVAYRRLAKY YHPDVYDGKG TLEEGETAEG RFIKIQAAYE LLMDTEKRRE YDMDNRVNPM 540 KASQAWMEWL MKKRKAFDQR GDMAIAAWAE QQQLEINLRA RRLSRSKVDP EEERKILEKE 600 KKASRELFNS TLKRHTLVLK KRDLMRKKAE EDKKKLITQL LAAEGLELDT EEEEEEEEET 660 AK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1 (By similarity). {ECO:0000250, ECO:0000269|PubMed:12953119}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00247 | DAP | Transfer from AT1G77920 | Download |
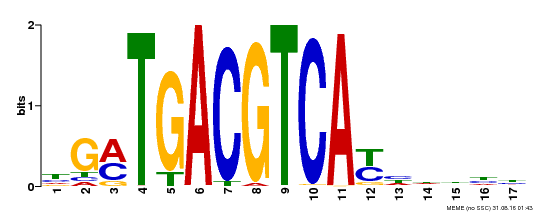
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353384 | 0.0 | AK353384.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-35-N08. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018443022.1 | 0.0 | PREDICTED: transcription factor TGA7-like | ||||
| Refseq | XP_018443023.1 | 0.0 | PREDICTED: transcription factor TGA7-like | ||||
| Swissprot | Q93ZE2 | 0.0 | TGA7_ARATH; Transcription factor TGA7 | ||||
| TrEMBL | A0A371FDF4 | 0.0 | A0A371FDF4_MUCPR; Transcription factor TGA3 (Fragment) | ||||
| STRING | XP_010416756.1 | 0.0 | (Camelina sativa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G77920.1 | 0.0 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



