 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC8230_p1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 800aa MW: 87569.5 Da PI: 5.8191 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.8 | 1.7e-12 | 395 | 441 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ L A+ YI++L+
RrC8230_p1 395 NHVEAERQRREKLNQRFYALRAVVPNV-----SKMDKASLLGDAIAYINELE 441
799***********************6.....5****************995 PP
| |||||||
| 2 | HLH | 20.4 | 9.4e-07 | 775 | 800 | 4 | 29 |
HHHHHHHHHHHHHHHHHHHHHCTSCC CS
HLH 4 ahnerErrRRdriNsafeeLrellPk 29
+h e+Er+RR+++N++f Lr ++P+
RrC8230_p1 775 NHVEAERQRREKLNQRFYALRAVVPN 800
799*********************95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 5.7E-57 | 44 | 228 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| SuperFamily | SSF47459 | 7.85E-17 | 390 | 456 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.66E-14 | 390 | 441 | No hit | No description |
| PROSITE profile | PS50888 | 17.111 | 391 | 440 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 5.6E-10 | 395 | 441 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 9.4E-17 | 395 | 461 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.5E-15 | 397 | 446 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF14215 | 3.4E-10 | 565 | 608 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| CDD | cd00083 | 8.89E-7 | 770 | 800 | No hit | No description |
| PROSITE profile | PS50888 | 11.458 | 771 | 800 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 6.67E-7 | 771 | 800 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.0E-6 | 775 | 800 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009269 | Biological Process | response to desiccation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009963 | Biological Process | positive regulation of flavonoid biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0043619 | Biological Process | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051090 | Biological Process | regulation of sequence-specific DNA binding transcription factor activity | ||||
| GO:2000068 | Biological Process | regulation of defense response to insect | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 800 aa Download sequence Send to blast |
MRNLWATDDN TSMMEAFLTS SDMSALWLQE TSATAPAGFN QETLQQRLQA LIEGTNEGWT 60 YAIFWQPSYD FSGDSVLGWG DGYYKGEEDK GKPKQRSTPL PFSTPADQEY RKKVLRELNS 120 LISGGGGPVD DAVDEEVTDT EWFFLVSMTQ SFASGAGLAG KAFSTGNVVW VSGSDQLSGS 180 SCERAKQGGV FGMQTIACIP SVNGVVELGS TEQIRRSSDL MNKVRLLFNF DVGTNWNLQG 240 ENNPSVWIND PIGASEPGNG QFSSKFENGG SSSTITGNPN HDSTLSPVHS QTLNPNFNNN 300 FSGEMNFSTS SKPRPGEILS FGNNGKRSSM NPDPIQFENK RKNSLGLIDD KVLSFGGGGE 360 TDHSDLEASV VKETPEKRPK KRGRKPANGK EEPLNHVEAE RQRREKLNQR FYALRAVVPN 420 VSKMDKASLL GDAIAYINEL ELNVNKTESE KIKIKNQLEE VKMELAGRKA STCGDLSSSS 480 SSSSATAPVG MEIEVKVIGW DAMIRVESSK RNYPAARLMS ALMDLELEVS HASMSVVNDL 540 MIQQATVKMG FRIYTQDQLR ASLISKIGGV FGMQTIACIP SVNGVVELGS TEQIRRSSDL 600 MNKVRLLFNF DVGTNWNLQG ENNPSVWIND PIGASEPGNG QFSSKFENGG SSSTITGNPN 660 HDSTLSPVHS QTLNPNFNNN FSGEMNFSTS SKPRPGEILS FGNNGKRSSM NPDPIQFENK 720 RKNSLGLIDD KVLSFGGGGE TDHSDLEASV VKETPEKRPK KRGRKPANGK EEPLNHVEAE 780 RQRREKLNQR FYALRAVVPN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rru_A | 2e-71 | 3 | 224 | 5 | 227 | Transcription factor MYC3 |
| 4ywc_A | 2e-71 | 3 | 224 | 5 | 227 | Transcription factor MYC3 |
| 4ywc_B | 2e-71 | 3 | 224 | 5 | 227 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 756 | 764 | KRPKKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator. Common transcription factor of light, abscisic acid (ABA), and jasmonic acid (JA) signaling pathways. With MYC3 and MYC4, controls additively subsets of JA-dependent responses. In cooperation with MYB2 is involved in the regulation of ABA-inducible genes under drought stress conditions. Can form complexes with all known glucosinolate-related MYBs to regulate glucosinolate biosynthesis. Binds to the MYC recognition site (5'-CACATG-3'), and to the G-box (5'-CACNTG-3') and Z-box (5'-ATACGTGT-3') of promoters. Binds directly to the promoters of the transcription factors PLETHORA1 (PLT1) and PLT2 and represses their expression. Negative regulator of blue light-mediated photomorphogenic growth and blue- and far-red-light regulated gene expression. Activates multiple TIFY/JAZ promoters. Positive regulator of lateral root formation. Regulates sesquiterpene biosynthesis. Subjected to proteasome-dependent proteolysis. The presence of the destruction element (DE) involved in turnover is required for the function to regulate gene transcription. {ECO:0000269|PubMed:12509522, ECO:0000269|PubMed:15208388, ECO:0000269|PubMed:15923349, ECO:0000269|PubMed:21321051, ECO:0000269|PubMed:21335373, ECO:0000269|PubMed:21954460, ECO:0000269|PubMed:22669881, ECO:0000269|PubMed:23142764, ECO:0000269|PubMed:23593022, ECO:0000269|PubMed:23943862, ECO:0000269|PubMed:9368419}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00084 | PBM | Transfer from AT1G32640 | Download |
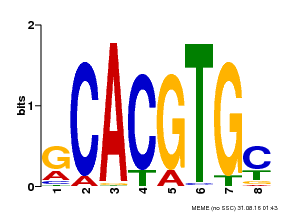
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Detected early after abscisic acid (ABA) treatment or after dehydration and high-salt stresses. Induced by UV treatment. Up-regulated by methyl jasmonate and herbivory. {ECO:0000269|PubMed:12679534, ECO:0000269|PubMed:15208388, ECO:0000269|PubMed:23593022, ECO:0000269|PubMed:23943862, ECO:0000269|PubMed:9368419}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018451259.1 | 0.0 | PREDICTED: transcription factor MYC2-like | ||||
| Swissprot | Q39204 | 0.0 | MYC2_ARATH; Transcription factor MYC2 | ||||
| TrEMBL | A0A0D3DKU7 | 0.0 | A0A0D3DKU7_BRAOL; Uncharacterized protein | ||||
| STRING | Bo8g028850.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1265 | 28 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32640.1 | 0.0 | bHLH family protein | ||||



