 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | RrC9921_p1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 963aa MW: 107601 Da PI: 5.3733 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 140.1 | 6.5e-44 | 10 | 96 | 32 | 118 |
CG-1 32 trpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylevk 118
++ gs++L++rk++ryfrkDG++w+kk+dgktv+E+he+LK g+v+vl+cyYah+++n++fqrr+yw+L+eel++iv+vhylevk
RrC9921_p1 10 NNELGGSVFLFDRKVLRYFRKDGHNWRKKRDGKTVKEAHERLKAGSVDVLHCYYAHGQDNENFQRRSYWMLQEELSHIVFVHYLEVK 96
555679******************************************************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 62.184 | 1 | 101 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.3E-48 | 1 | 96 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.6E-36 | 10 | 95 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.4E-16 | 425 | 507 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 4.37E-11 | 584 | 714 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 3.4E-15 | 585 | 717 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 17.237 | 588 | 714 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.55E-15 | 600 | 717 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.11 | 655 | 687 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.65E-8 | 808 | 862 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.92 | 811 | 833 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.029 | 813 | 841 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0038 | 814 | 832 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0011 | 834 | 856 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.468 | 835 | 859 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.1E-4 | 837 | 856 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 963 aa Download sequence Send to blast |
MAEARRFSPN NELGGSVFLF DRKVLRYFRK DGHNWRKKRD GKTVKEAHER LKAGSVDVLH 60 CYYAHGQDNE NFQRRSYWML QEELSHIVFV HYLEVKGSRV STSYNRMQRT EDSAPSSQET 120 GEVYTSERNG YASGSINQYD HSNHSQATDS ASVNGVHTPE LEDAESAYNQ QGSSILYSHQ 180 ALQQPPATSF DPYYQMSLTP RDSYQKDLRT IPSSTMVEKG RTINGPVLTN SIKTKKSIDS 240 QTWEEILGNC GSGGGEGLPM QPHREHEVLD QMLESYSFTM QDFASLQESM VKSQNQELNS 300 GLTSDHSMWF QGQAVDIEPN ALSNLASSEK APYLSTMKQH LLDGALGEEG LKKMDSFNRW 360 MSKELGELGD VGVTPDANES FTHSSSTAYW EEVESEDVSN VHNSRRDLDG YVMSPSLSKE 420 QLFSIIDFAP NWTYVGCEVK VLVSGKFLKT AETREWSCMF GQTEVPADII ANGILECVAP 480 MHEAGRVPFY VTCSNRLACS EVREFEYKVL ESQVFDRETD DSTACNSIES LEARFVKLLC 540 SKSDCPNTSL SGNDSDLSQV SEKISLLLFE NDDGLDQMLM NEISQENMKN NLLQEALKES 600 LHSWLLQKIA EGGKGPNVLD EGGQGILHFA AALGYNWALE PTIVAGVSVD FRDVNGWTAL 660 HWAAFFGREL IIGSLIALGA SPGTLTDPNP DFPSGSTPSD LAYANGYKGI AGYLSEYALR 720 AHVSLLSLNE KNAAVETAPS PSSSALTDSL TAVRNASQAA ARIHQVFRAQ SFQKKQMKEL 780 GDRKLGMSEE RAISMLAPKT HKQGRAHSDD SVQAAAIRIQ NKFRGYKGRK DYLITRQRII 840 KIQAHVRGYQ VRKNYRKIIW SVGILEKVIL RWRRKGAGLR GFKSDALVTK MQDGTEKEED 900 DDFFKQGRKQ TEERLEKALA RVEKALANSE EAACFDDDLI DIEALLGDDD TLMMPMPSTL 960 WNT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
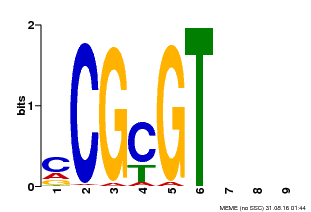
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018483642.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 3 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A3P6CXK7 | 0.0 | A0A3P6CXK7_BRACM; Uncharacterized protein | ||||
| STRING | Bra030248.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8601 | 25 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||



