 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_00060.1_g00037.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 402aa MW: 46002.6 Da PI: 7.5175 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11 | 0.0014 | 33 | 55 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y C++Cg s s k++ ++Hi +H
Rsa1.0_00060.1_g00037.1 33 YLCQYCGISRSKKYLITSHIESH 55
56*******************96 PP
| |||||||
| 2 | zf-C2H2 | 20.2 | 1.6e-06 | 81 | 103 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C+ Cg F+ + +Lk+H+++H
Rsa1.0_00060.1_g00037.1 81 HTCQECGAEFKKPAHLKQHMQSH 103
79*******************99 PP
| |||||||
| 3 | zf-C2H2 | 17.2 | 1.4e-05 | 127 | 151 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ C dC+ s++rk++L+rH+ tH
Rsa1.0_00060.1_g00037.1 127 FACYvdDCTASYRRKDHLNRHLLTH 151
678888*****************99 PP
| |||||||
| 4 | zf-C2H2 | 19.9 | 1.9e-06 | 205 | 228 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirtH 23
+C+ Cgk F+ +s+L++H +H
Rsa1.0_00060.1_g00037.1 205 VCKevGCGKAFKYPSQLQKHEDSH 228
79999***************9888 PP
| |||||||
| 5 | zf-C2H2 | 20.2 | 1.6e-06 | 295 | 320 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kC+ C+ +Fs +snL++H++ H
Rsa1.0_00060.1_g00037.1 295 FKCEveGCSSTFSKPSNLQKHLKAvH 320
89********************9877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 1.47E-6 | 31 | 103 | No hit | No description |
| SMART | SM00355 | 26 | 33 | 55 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 9.7E-6 | 79 | 104 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0015 | 81 | 103 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.988 | 81 | 108 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 83 | 103 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.6E-9 | 120 | 153 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.23E-6 | 123 | 153 | No hit | No description |
| PROSITE profile | PS50157 | 9.66 | 127 | 156 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.14 | 127 | 151 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 129 | 151 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 8.8E-4 | 154 | 182 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.28E-5 | 154 | 182 | No hit | No description |
| PROSITE profile | PS50157 | 9.91 | 156 | 186 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.62 | 156 | 181 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 158 | 181 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 5.4E-6 | 199 | 228 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 9.1E-5 | 200 | 229 | No hit | No description |
| PROSITE profile | PS50157 | 10.471 | 204 | 233 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0055 | 204 | 228 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 206 | 228 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.2 | 236 | 261 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 238 | 261 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 25 | 264 | 285 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 6.86E-9 | 275 | 333 | No hit | No description |
| SMART | SM00355 | 3.6E-4 | 295 | 320 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 8.5E-8 | 295 | 320 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 12.84 | 295 | 325 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 297 | 320 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 8.9E-5 | 321 | 347 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 4.9 | 326 | 352 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 328 | 352 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 402 aa Download sequence Send to blast |
MNPSPCVDTQ REMSEEEANV DVNPTKTKDI RNYLCQYCGI SRSKKYLITS HIESHHKLEV 60 EEERNEEACE DDEEEISGKK HTCQECGAEF KKPAHLKQHM QSHSLEYQAG RPPEKLPASK 120 PPSRRPFACY VDDCTASYRR KDHLNRHLLT HKGKLFKCPV ENCKSEFSVH GNISKHVKKF 180 HTDSDSNKDE DSQPTESSSG QKKLVCKEVG CGKAFKYPSQ LQKHEDSHVK LDSVEAFCSE 240 IGCMKYFTNE ECLKAHIRSC HQHVNCEICG SKHLKKNIKR HLRTHDEDSS SPGEFKCEVE 300 GCSSTFSKPS NLQKHLKAVH EDIRPFVCGF PGCGMRFAYK HVRNNHENSG SHIYTCGDFV 360 EADEDFTSRP RGGLKRKQVT AEMLVRKRVM PPQFDSEEHE TC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wjq_D | 1e-12 | 33 | 355 | 9 | 284 | Zinc finger protein 568 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
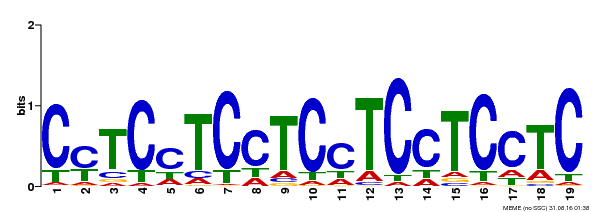
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_00060.1_g00037.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ630478 | 0.0 | AJ630478.1 Arabidopsis thaliana mRNA for hypothetical protein, clone At1g72050D. | |||
| GenBank | AY054225 | 0.0 | AY054225.1 Arabidopsis thaliana At1g72050/F28P5_6 mRNA, complete cds. | |||
| GenBank | AY066042 | 0.0 | AY066042.1 Arabidopsis thaliana At1g72050/F28P5_6 mRNA, complete cds. | |||
| GenBank | AY186610 | 0.0 | AY186610.1 Arabidopsis thaliana transcription factor IIIA (TFIIIA) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018446244.1 | 0.0 | PREDICTED: transcription factor IIIA-like | ||||
| Swissprot | Q84MZ4 | 0.0 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A3P6BJ38 | 0.0 | A0A3P6BJ38_BRACM; Uncharacterized protein (Fragment) | ||||
| STRING | Bo6g092790.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4714 | 28 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 0.0 | transcription factor IIIA | ||||



