 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_00098.1_g00023.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 390aa MW: 42760.7 Da PI: 7.9705 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 62.7 | 7.9e-20 | 181 | 230 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrd.psengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+y+GVr+++ +g+W+AeIrd + + +r++lg+f+ ae Aa+a+++a+++++g
Rsa1.0_00098.1_g00023.1 181 RYRGVRQRP-WGKWAAEIRDpF-----KaARVWLGTFDNAESAARAYDEAALRFRG 230
7********.**********66.....56*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 3.0E-13 | 181 | 230 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 25.106 | 181 | 238 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.5E-32 | 181 | 239 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 8.4E-36 | 181 | 244 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 5.36E-22 | 181 | 239 | IPR016177 | DNA-binding domain |
| PRINTS | PR00367 | 5.1E-11 | 182 | 193 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.71E-26 | 189 | 240 | No hit | No description |
| PRINTS | PR00367 | 5.1E-11 | 204 | 220 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006970 | Biological Process | response to osmotic stress | ||||
| GO:0009749 | Biological Process | response to glucose | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 390 aa Download sequence Send to blast |
MCTIKVANQE GNVGKKAEFT TREDDHGTLS DIDQWLYLFA AEDDHHLHRD NALIPPSSSS 60 LVSYSREMEM SAIVSALTHV VAGNVPTHQD GGGGERTSNS SSSSGHKRRR EVDEDGGGGK 120 AVKAPNTLTV DQYFSSGSST SKVREGSSNL SGGSGPTYEY MTTTNGNTET SSSSGDQPRR 180 RYRGVRQRPW GKWAAEIRDP FKAARVWLGT FDNAESAARA YDEAALRFRG NKAKLNFPEN 240 VKLVKPAATQ SVPPTAVQRP TQLRNSGSTS TLLPIRPASD QIVHSQPLMQ PYNLSYSEMA 300 RQQQFQHHHH HHQQSRDLYD QISFPLSFGH SRGSTMLSTS SSSSHSRPLF SPAAVQSTPE 360 SVSETGYLQD VQWSPDKTSD NNNYTNSPST |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 1e-22 | 180 | 241 | 1 | 63 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Negative regulator of the abscisic acid (ABA) signaling pathway involved in seed germination and in responses to stress conditions. Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250, ECO:0000269|PubMed:16227468}. | |||||
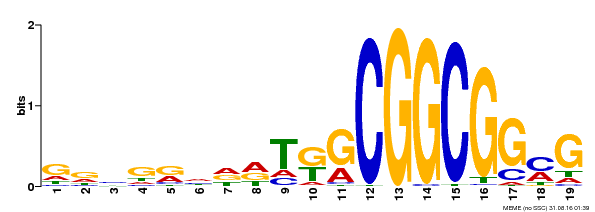
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00584 | DAP | Transfer from AT5G64750 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_00098.1_g00023.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (ABA) and stresses, including cold, drought and salt (PubMed:16227468, PubMed:26961720). Triggered by YY1 (PubMed:26961720). {ECO:0000269|PubMed:16227468, ECO:0000269|PubMed:26961720}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018489015.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor ABR1 | ||||
| Swissprot | Q9FGF8 | 1e-161 | ABR1_ARATH; Ethylene-responsive transcription factor ABR1 | ||||
| TrEMBL | M4E6C1 | 0.0 | M4E6C1_BRARP; Uncharacterized protein | ||||
| STRING | Bra024325.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9118 | 21 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64750.1 | 1e-132 | ERF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



