 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_00145.1_g00029.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 312aa MW: 34749.5 Da PI: 4.8893 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 34.4 | 5.1e-11 | 61 | 102 | 3 | 46 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
++T+eE++l+++ ++ +G++ W +Ia++++ g+t++++k w++
Rsa1.0_00145.1_g00029.1 61 SFTPEEEDLIIKMHAAMGSR-WPLIAQHLP-GKTEEEVKMVWNT 102
79******************.*********.********88876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.22E-12 | 41 | 112 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.732 | 54 | 108 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-16 | 54 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.7E-11 | 58 | 106 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.70E-9 | 61 | 104 | No hit | No description |
| Pfam | PF00249 | 3.2E-10 | 61 | 102 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 312 aa Download sequence Send to blast |
MVRSCSSKSK NPWTDEDNAP QKFAFASASK NGSTPKKIGL RRCGKSCRVR KTENAGAKHE 60 SFTPEEEDLI IKMHAAMGSR WPLIAQHLPG KTEEEVKMVW NTKLKKKLSQ MGIDHVTHRP 120 FSHVLAEYGN INGGGTLNPN PMNQTGSLGP NPSFNEDSHE QQQQQPDDSG DLMFHLQAIK 180 LMTESSNQVK PESTFMYASS SSSNSSPPLF SSTCSTIAQE SSEVNFTWSD FLLDQETFSE 240 NQQQNYPDQE LDNLFGNEFS EAEAVTTMAN TATPASQMEE ESLSNGFVES IIAKEKELFL 300 GFPSYLEQPF HF |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00148 | DAP | Transfer from AT1G18960 | Download |
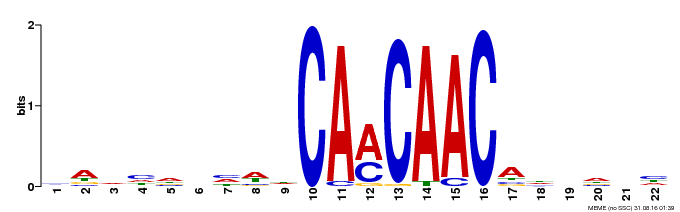
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_00145.1_g00029.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018449663.1 | 0.0 | PREDICTED: transcription factor MYB35 | ||||
| TrEMBL | A0A0D3DQA7 | 0.0 | A0A0D3DQA7_BRAOL; Uncharacterized protein | ||||
| STRING | Bo8g068210.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM14822 | 15 | 18 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G18960.1 | 1e-167 | MYB_related family protein | ||||



