 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_00191.1_g00036.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 599aa MW: 65357.4 Da PI: 4.9934 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.5 | 1e-12 | 428 | 473 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ L A+ YI++L
Rsa1.0_00191.1_g00036.1 428 NHVEAERQRREKLNQRFYSLRAVVPNV-----SKMDKASLLGDAISYINEL 473
799***********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 1.2E-53 | 63 | 258 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.282 | 424 | 473 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.31E-14 | 427 | 478 | No hit | No description |
| SuperFamily | SSF47459 | 4.06E-18 | 427 | 498 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.5E-10 | 428 | 473 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.0E-18 | 428 | 494 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.5E-16 | 430 | 479 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006952 | Biological Process | defense response | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043425 | Molecular Function | bHLH transcription factor binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 599 aa Download sequence Send to blast |
MSSTDVQVTD HHLNQSTNGT NLWSTTEDDA SVMEPFIGSE HSSLWPQPPL TPPPPPHVTE 60 DTLQQRLQAL IEGARESWTY AVFWQLSHGF AGEDSTTAAA LLSWGDGYYK GEEEERKKRK 120 PNNPVSAAEQ EHRKRVIREL NSLISGGGGG TVSGSSSDEA GDEDVTDAEW FFLVSMTQSF 180 FASDGSGLPG RAFSNSQTIW LSGSNALAGS SCERARQGQV YGLETMVCVP TENGVVELGS 240 LEVIHQSSDL VDKVNGFFSF NGGGGGVSGS WAFNLNPDQG ENDPATTWIN EPDVTGIEPV 300 LGVNTGNNNN STSSNSDSQT ASKLCNGSSV EHPKQQQQHQ NPQISSSGFV EGDSKKKKRC 360 LVSDKEEEML SFTSVLPLAT KTNDSDHSDL EASVAKGQKS GGRTVVEPEK KPRKRGRKPA 420 NGREESLNHV EAERQRREKL NQRFYSLRAV VPNVSKMDKA SLLGDAISYI NELKAKLEKA 480 ECDKEELKKK IDGMSQEVAS GNVKSSVKDQ KCVDQDSGVS VEVEIDVKII GWDAMIRIQC 540 GKKNHPGAKF MEALRELELE VNHASLSVVN EFMIQQATVK MGNQFFTQDK LKAALMERV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rru_A | 1e-81 | 25 | 254 | 8 | 227 | Transcription factor MYC3 |
| 4ywc_A | 1e-81 | 25 | 254 | 8 | 227 | Transcription factor MYC3 |
| 4ywc_B | 1e-81 | 25 | 254 | 8 | 227 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 409 | 417 | KKPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in jasmonic acid (JA) gene regulation. With MYC2 and MYC3, controls additively subsets of JA-dependent responses. Can form complexes with all known glucosinolate-related MYBs to regulate glucosinolate biosynthesis. Binds to the G-box (5'-CACGTG-3') of promoters. Activates multiple TIFY/JAZ promoters. {ECO:0000269|PubMed:21321051, ECO:0000269|PubMed:21335373, ECO:0000269|PubMed:23943862}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00086 | PBM | Transfer from AT4G17880 | Download |
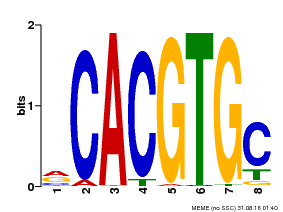
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_00191.1_g00036.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. Not induced by jasmonic acid. {ECO:0000269|PubMed:12679534, ECO:0000269|PubMed:21335373}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC183492 | 0.0 | AC183492.1 Brassica oleracea Contig G, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018471791.1 | 0.0 | PREDICTED: transcription factor MYC4 | ||||
| Swissprot | O49687 | 0.0 | MYC4_ARATH; Transcription factor MYC4 | ||||
| TrEMBL | A0A078I392 | 0.0 | A0A078I392_BRANA; BnaC01g10420D protein | ||||
| STRING | Bo1g017990.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1265 | 28 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17880.1 | 0.0 | bHLH family protein | ||||



