 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_00730.1_g00008.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 604aa MW: 69027.7 Da PI: 7.1207 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 48.2 | 2.8e-15 | 94 | 171 | 2 | 82 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkr..tsessstcpyf 82
W+ +evlaL+++r+ +e+++ + We s+k++e gf+rsp++Ckek+e+ ++ry + + + + + +++ ++++f
Rsa1.0_00730.1_g00008.1 94 WCSDEVLALLRFRSTVENWFPEF-----TWELTSRKLAEVGFKRSPQECKEKFEEEERRYFNSNSANDHHisNYNNKGNYRMF 171
********************998.....9**********************************99999884223344455555 PP
| |||||||
| 2 | trihelix | 98.4 | 6e-31 | 447 | 543 | 1 | 86 |
trihelix 1 rWtkqevlaLiearr...........emeerlrrgk.........lkkplWeevskkmrergferspkqCkekwenlnkrykkike 66
rW+++evlaLi++rr + ++ lWe++skkm e g++rs+k+Ckekwen+nk++kk+k+
Rsa1.0_00730.1_g00008.1 447 RWPRDEVLALINIRRsistmnddehhK--------DgislsssspKAVRLWERISKKMLESGYKRSAKRCKEKWENINKYFKKTKD 524
8**************666554333322........0344444455899************************************** PP
trihelix 67 gekkrtsessstcpyfdqle 86
+kkr + +s+tcpyf+ql+
Rsa1.0_00730.1_g00008.1 525 VNKKR-PLDSRTCPYFHQLT 543
****8.9***********98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF13837 | 3.1E-10 | 92 | 156 | No hit | No description |
| PROSITE profile | PS50090 | 5.482 | 93 | 145 | IPR017877 | Myb-like domain |
| CDD | cd12203 | 6.85E-24 | 446 | 523 | No hit | No description |
| Pfam | PF13837 | 6.0E-20 | 446 | 544 | No hit | No description |
| PROSITE profile | PS50090 | 6.864 | 447 | 516 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 604 aa Download sequence Send to blast |
MFDGGVPEQI QQFIASPQPP PPPHQPATES SLPFPVSFAS FNSNHQAQHM LSLDSRKIIH 60 HHHHDIKDGG VTSEWIGHTD HDGEDHRHFH HHPWCSDEVL ALLRFRSTVE NWFPEFTWEL 120 TSRKLAEVGF KRSPQECKEK FEEEERRYFN SNSANDHHIS NYNNKGNYRM FSEAEEFYHH 180 GHNDDEHVPS EVGDNQNKSN NSLEGKGNVE ETGQDLLDDK RDHQGQVKES SMGNKVTTVD 240 NVGNHEDDGK SSSSSLNLKM IMRDKKKRKR KKKKERFVVL TGFFEGVVSN MIAQQEEMHK 300 KLLGDMVKKE EEKVAREEAW KKQEMERLNK EVEIRANEQA MASDRNTSII KFISKFTDHD 360 SHDDGNYKVH SPSPSQDLSS LVLPKTQGIT ECQTSSALLP RTLTSDYPIS LQTNDIPLDP 420 ISTKTLKTKT QNIKPPKSDD KSDISKRWPR DEVLALINIR RSISTMNDDE HHKDGISLSS 480 SSPKAVRLWE RISKKMLESG YKRSAKRCKE KWENINKYFK KTKDVNKKRP LDSRTCPYFH 540 QLTALYSQPF TGTTTTATNT STGELETRVG SGDHDDIPAA MHLDANGASK KNSVPFSGFD 600 LAFE |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 265 | 270 | KKRKRK |
| 2 | 265 | 272 | KKRKRKKK |
| 3 | 265 | 273 | KKRKRKKKK |
| 4 | 266 | 274 | KRKRKKKKE |
| 5 | 267 | 271 | RKRKK |
| 6 | 267 | 273 | RKRKKKK |
| 7 | 267 | 274 | RKRKKKKE |
| 8 | 269 | 274 | RKKKKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specific DNA sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
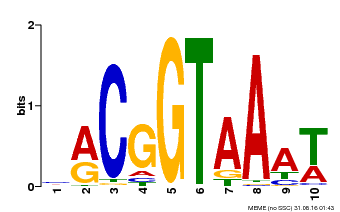
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_00730.1_g00008.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013677968.1 | 0.0 | trihelix transcription factor GTL2 | ||||
| Swissprot | Q8H181 | 0.0 | GTL2_ARATH; Trihelix transcription factor GTL2 | ||||
| TrEMBL | A0A3P6E5R7 | 0.0 | A0A3P6E5R7_BRAOL; Uncharacterized protein | ||||
| STRING | Bo2g152980.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8268 | 28 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 0.0 | Trihelix family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



