 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_00861.1_g00002.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 435aa MW: 48303 Da PI: 8.4579 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 125.1 | 2.3e-39 | 106 | 166 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++k+l+cprC+s++tkfCyynny+++qPr+fCkaC+ryWt+GG++rnvPvG+grrk+k+ss
Rsa1.0_00861.1_g00002.1 106 PTKILPCPRCKSMDTKFCYYNNYNINQPRHFCKACQRYWTAGGTMRNVPVGAGRRKHKSSS 166
67899*****************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 3.0E-28 | 103 | 158 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 6.3E-32 | 108 | 164 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.435 | 110 | 164 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 112 | 148 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 435 aa Download sequence Send to blast |
MMMESRDPAI KLFGMKIPFP AVFEQPLTVA VVEEEHTGGD DKSLEKVTTE QVTPELSEKK 60 NNCNNNNNLN DSKPETGDKE EEATSTDDQT ESDQQTTEDG KTLKKPTKIL PCPRCKSMDT 120 KFCYYNNYNI NQPRHFCKAC QRYWTAGGTM RNVPVGAGRR KHKSSSSHYR HITISEALQA 180 ARLDPGLQSN TRVLSFGLQA HQQHAAPITP VMKLQGDQKV QNGARNGLAD QRVENGDDCS 240 SGSSVTTTSV DETRAQNCRV VEPQMNSNNN MNGYACIPGV PWPYTWNPAM PPPGFYPPPG 300 YPMPFYPYWT IPMLPPNQSS SPISQKGWSP NSPTLGKHSR DEEPSRKGDE TERKQRNGRV 360 IVPKTLRIDD PDEAAKSSIW TTLGIKNEGS RLGSKGGGMF KGFDQKTKKS NNENPPLLSA 420 NPAALSRSLN FQERV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Regulates a photoperiodic flowering response. Transcriptional repressor of 'CONSTANS' expression. {ECO:0000250, ECO:0000269|PubMed:19619493}. | |||||
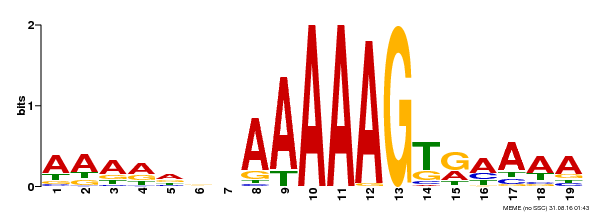
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00394 | DAP | Transfer from AT3G47500 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_00861.1_g00002.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Highly expressed at the beginning of the light period, then decreases, reaching a minimum between 16 and 29 hours after dawn before rising again at the end of the day. {ECO:0000269|PubMed:19619493}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | GU451312 | 0.0 | GU451312.1 Brassica napus putative dof zinc finger protein (dof1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018441626.1 | 0.0 | PREDICTED: cyclic dof factor 3 | ||||
| Swissprot | Q8LFV3 | 0.0 | CDF3_ARATH; Cyclic dof factor 3 | ||||
| TrEMBL | A0A3P5YH86 | 0.0 | A0A3P5YH86_BRACM; Uncharacterized protein | ||||
| STRING | Bo3g134590.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5441 | 28 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G47500.1 | 0.0 | cycling DOF factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



