 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_00944.1_g00011.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 277aa MW: 31070.2 Da PI: 9.8951 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 61.9 | 2.8e-19 | 1 | 113 | 1 | 95 |
GAGA_bind 1 mdddgsre..rnkg.yye.................paaslkenlglqlmssiaerdakirernlalsekkaavaerdmaflqrdka 66
m++ g+++ r k+ y++ ++++l +++++ms++aerd++++ern a+++kk+a+a+rd+a+ qrd a
Rsa1.0_00944.1_g00011.1 1 MESGGQYDngRYKPdYFKgtppsvwnimpqhqikeQHNALV--MNKKIMSILAERDSALKERNEAVAAKKEALAARDEALEQRDRA 84
888999888999999999**********9887764446666..99***************************************** PP
GAGA_bind 67 laernkalverdnkllalllvenslasal 95
l+er++a++er+ +l al + en+l+++
Rsa1.0_00944.1_g00011.1 85 LSERDNAIMERETALNALHYPENNLNHHI 113
***********************998764 PP
| |||||||
| 2 | GAGA_bind | 251.4 | 5.6e-77 | 134 | 277 | 156 | 301 |
GAGA_bind 156 akkpkekkakkkkkksekskkkvkkesaderskaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSv 241
++++k +k+kk+ k++++ ++ v++++ ++ +++++ d++ln ++De+++PvPvC+CtG+ rqCYkWG+GGWqS+CCttt+S+
Rsa1.0_00944.1_g00011.1 134 PADKKPTKRKKESKQGKDLNRLVASPG--KKCRKDWDVNDVGLNLLAFDETSMPVPVCTCTGTARQCYKWGSGGWQSSCCTTTLSQ 217
455666677777777888888899998..57******************************************************* PP
GAGA_bind 242 yPLPvstkrrgaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
+PLP+++++r++R++grKmS+++f++lL++La+eG+dls pvDLkd+WA+HGtn+++ti+
Rsa1.0_00944.1_g00011.1 218 HPLPQMPNKRHSRMGGRKMSGNVFSRLLSRLAGEGHDLSSPVDLKDYWARHGTNRYITIK 277
***********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 2.2E-139 | 1 | 277 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 2.8E-83 | 18 | 277 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 277 aa Download sequence Send to blast |
MESGGQYDNG RYKPDYFKGT PPSVWNIMPQ HQIKEQHNAL VMNKKIMSIL AERDSALKER 60 NEAVAAKKEA LAARDEALEQ RDRALSERDN AIMERETALN ALHYPENNLN HHILSRGGNE 120 GTHLPNPSPP STIPADKKPT KRKKESKQGK DLNRLVASPG KKCRKDWDVN DVGLNLLAFD 180 ETSMPVPVCT CTGTARQCYK WGSGGWQSSC CTTTLSQHPL PQMPNKRHSR MGGRKMSGNV 240 FSRLLSRLAG EGHDLSSPVD LKDYWARHGT NRYITIK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000269|PubMed:14731261}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00480 | DAP | Transfer from AT4G38910 | Download |
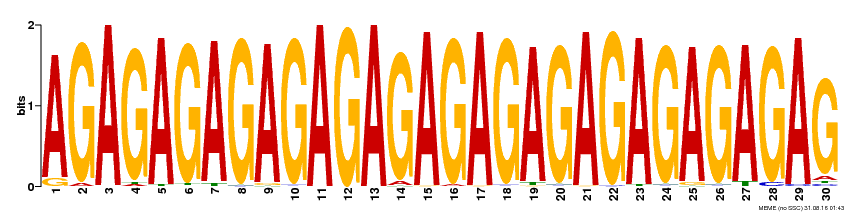
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_00944.1_g00011.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018447695.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE5-like | ||||
| Refseq | XP_018447854.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE5-like | ||||
| Swissprot | F4JUI3 | 1e-158 | BPC5_ARATH; Protein BASIC PENTACYSTEINE5 | ||||
| TrEMBL | A0A3P6C5P1 | 1e-176 | A0A3P6C5P1_BRACM; Uncharacterized protein | ||||
| STRING | XP_006411681.1 | 1e-173 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4090 | 26 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G38910.2 | 1e-146 | basic pentacysteine 5 | ||||



