 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_00949.1_g00010.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 580aa MW: 64234.1 Da PI: 6.8993 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.4 | 1.1e-12 | 422 | 467 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr+++P+ + K++Ka+ L Av YI++L
Rsa1.0_00949.1_g00010.1 422 NHVEAERQRREKLNQRFYALRSVVPNiS------KMDKASLLGDAVSYINEL 467
799***********************66......***************988 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 7.8E-50 | 49 | 239 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.136 | 418 | 467 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.09E-18 | 421 | 486 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.19E-14 | 421 | 472 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-17 | 422 | 481 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.0E-10 | 422 | 467 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.6E-16 | 424 | 473 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 580 aa Download sequence Send to blast |
MSSNNNRVWN EDDKAIVASL LGKKALDYLL SNSVPNANLL MTVGSDDNLQ NKLSDLVETP 60 NSSNFSWNYA IFWQVSRSKS GDLVLCWGDG SCREPKDGEK SEMMRMLSMG REEETHQTLR 120 KRVLQKLHAL FGGLDDEEDN CALVLDRVTD TEMFLLASMY FSFPRGQGGP GKCFHSSHPV 180 WLSDLVNSGS DYCVRSFLAK SAGIQTVVLV PTDIGVVELG STSCLPHSDD SISSIRSSFS 240 SPPPPPPPVR VVLPVVVADD NSSSKIFGKD LHTSPAFLHH HHQQQQHNRQ FREKLTVRKM 300 DDRVPKRLDG NRFMFSNPST TPTWIPPPRT NNNSVTAEAP SREDFKFLPL QQSSSSSHRL 360 LPPAQMQIDF SGASSRASET HSDGGGGADW ADLVGGADES GGDNRPRKRG RRPANGRVEA 420 LNHVEAERQR REKLNQRFYA LRSVVPNISK MDKASLLGDA VSYINELYAK LKVMEAERER 480 LGYSSNPHLC LEQPVINVQT AGEDVAVTVS CPLDSHPASR IFHAFEEAKV EVINSKLEVP 540 QDLVLHTFVI KSEEVTKEKL ISALSREQSS SVQSRTSSGR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 3e-25 | 416 | 481 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_B | 3e-25 | 416 | 481 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_E | 3e-25 | 416 | 481 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_F | 3e-25 | 416 | 481 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_G | 3e-25 | 416 | 481 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_I | 3e-25 | 416 | 481 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_M | 3e-25 | 416 | 481 | 2 | 65 | Transcription factor MYC2 |
| 5gnj_N | 3e-25 | 416 | 481 | 2 | 65 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
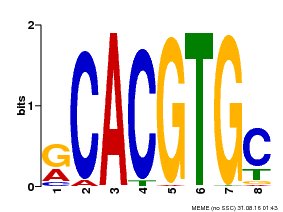
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_00949.1_g00010.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018441162.1 | 0.0 | PREDICTED: transcription factor bHLH13 | ||||
| Swissprot | Q9LNJ5 | 0.0 | BH013_ARATH; Transcription factor bHLH13 | ||||
| TrEMBL | A0A397XPQ5 | 0.0 | A0A397XPQ5_BRACM; Uncharacterized protein | ||||
| STRING | Bra033273.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3758 | 27 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 0.0 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



