 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_01020.1_g00009.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 288aa MW: 33157.6 Da PI: 6.9894 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 45.5 | 1.7e-14 | 14 | 62 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT.-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGgg.tWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEd +l+ +++ G+g +W + ++++g++R++k+c++rw++yl
Rsa1.0_01020.1_g00009.1 14 KGPWSPEEDAKLKSYIETSGTGgNWIALPHKIGLKRCGKSCRLRWLNYL 62
79*********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 38 | 3.8e-12 | 69 | 111 | 2 | 46 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
g +++eE+ ++ ++ G++ W+ Ia+ ++ gRt++++k++w++
Rsa1.0_01020.1_g00009.1 69 GGFSEEEENIICNLYLTIGSR-WSIIAAQLP-GRTDNDIKNYWNT 111
569******************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.464 | 9 | 62 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.56E-25 | 11 | 109 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.9E-11 | 13 | 64 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.0E-14 | 14 | 62 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-23 | 15 | 69 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.48E-7 | 16 | 62 | No hit | No description |
| PROSITE profile | PS51294 | 20.546 | 63 | 117 | IPR017930 | Myb domain |
| SMART | SM00717 | 8.1E-11 | 67 | 115 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.2E-11 | 69 | 111 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-23 | 70 | 118 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.50E-7 | 71 | 113 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 288 aa Download sequence Send to blast |
MGRAPCCDKT NVKKGPWSPE EDAKLKSYIE TSGTGGNWIA LPHKIGLKRC GKSCRLRWLN 60 YLRPNIKHGG FSEEEENIIC NLYLTIGSRW SIIAAQLPGR TDNDIKNYWN TRLKKKLIIK 120 QRKELQEGCM EQQQMMVMMK RQQEQQQIQT TFTMRQDQTI FTWPLQQLPY HHHHDRVPNL 180 VMNSLGDQED IKPAITKNMA KIEDQELERT NALDHLSFSQ LLLDHNNNLG EGLSMNSILS 240 TNTSNNHQWF GNVIDEDIDL FSGATSTSAD QDTIRWEDIS SLVCPDSK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-23 | 14 | 117 | 7 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (By similarity). Positively regulates axillary meristems (AMs) formation and development, especially during inflorescence. {ECO:0000250, ECO:0000269|PubMed:16461581}. | |||||
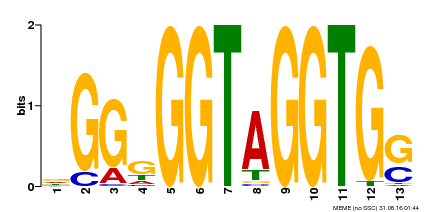
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00089 | SELEX | Transfer from AT3G49690 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_01020.1_g00009.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY519597 | 1e-168 | AY519597.1 Arabidopsis thaliana MYB transcription factor (At3g49690) mRNA, complete cds. | |||
| GenBank | BT029224 | 1e-168 | BT029224.1 Arabidopsis thaliana At3g49690 mRNA, complete cds. | |||
| GenBank | Y14209 | 1e-168 | Y14209.1 A.thaliana mRNA for AtMYB84 R2R3-MYB transcription factor. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018468640.1 | 0.0 | PREDICTED: transcription factor RAX3 | ||||
| Swissprot | Q9M2Y9 | 1e-153 | RAX3_ARATH; Transcription factor RAX3 | ||||
| TrEMBL | A0A0D3DRN6 | 1e-175 | A0A0D3DRN6_BRAOL; Uncharacterized protein | ||||
| STRING | Bo8g078940.1 | 1e-176 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2941 | 25 | 68 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G49690.1 | 1e-137 | myb domain protein 84 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



