 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_01036.1_g00011.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 569aa MW: 65146.6 Da PI: 8.087 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 96.5 | 2.3e-30 | 42 | 126 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW++ e+laL+++r+em++++r++ lk+plWee+s+km+e g++rs+k+Ckek+en+ k++k++keg+ ++++++ t+++f++le
Rsa1.0_01036.1_g00011.1 42 RWPRPETLALLRIRSEMDKSFRDSTLKAPLWEEISRKMMELGYKRSAKKCKEKFENVYKYHKRTKEGRTGKSEGK--TYRFFEELE 125
8********************************************************************975544..6******98 PP
trihelix 87 a 87
a
Rsa1.0_01036.1_g00011.1 126 A 126
5 PP
| |||||||
| 2 | trihelix | 108.8 | 3.4e-34 | 391 | 476 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+k ev aLi++r+++e +++++ k+plWee+s+ mr+ g++rs+k+Ckekwen+nk++kk+ke++kkr + +s+tcpyf+qle
Rsa1.0_01036.1_g00011.1 391 RWPKTEVEALIRIRKNLEANYQENGTKGPLWEEISAGMRRLGYNRSAKRCKEKWENINKYFKKVKESNKKR-PLDSKTCPYFHQLE 475
8*********************************************************************8.9************8 PP
trihelix 87 a 87
a
Rsa1.0_01036.1_g00011.1 476 A 476
5 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 0.11 | 39 | 101 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 2.77E-24 | 41 | 106 | No hit | No description |
| PROSITE profile | PS50090 | 7.015 | 41 | 99 | IPR017877 | Myb-like domain |
| Pfam | PF13837 | 2.2E-20 | 41 | 126 | No hit | No description |
| SMART | SM00717 | 9.2E-4 | 388 | 450 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS50090 | 7.491 | 390 | 448 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-4 | 390 | 447 | IPR009057 | Homeodomain-like |
| Pfam | PF13837 | 7.0E-24 | 390 | 477 | No hit | No description |
| CDD | cd12203 | 1.37E-25 | 390 | 455 | No hit | No description |
| SuperFamily | SSF46689 | 6.22E-5 | 391 | 482 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 569 aa Download sequence Send to blast |
MSGNSLGLLD SSGGGVGGSA EEEKDMKMEE TNEVGGGGGG SRWPRPETLA LLRIRSEMDK 60 SFRDSTLKAP LWEEISRKMM ELGYKRSAKK CKEKFENVYK YHKRTKEGRT GKSEGKTYRF 120 FEELEAFESQ PAKSPAATTT TIATTPLIPW ISSTTPSTEK TSSPLKHQQH HQVSVKPIAT 180 SPTFLTKQPS STTPFPFYSN NHTTTLDTGH KPTSNDLVNN VSSLNLFSSS TSSSTASDEE 240 DDQEKRSRKK RKYWKGLFTK LTKELMEKQE KMQKRFLESL ENREKERISR EEAWRVQEVA 300 RISREYETLV NERSNAAAKD AAIISFLHKI SGGQQQQQPQ QQPQQNHKVP QRKQYQSEQH 360 SITFESKEPR PVLLETTMKM GNHSLSPSSS RWPKTEVEAL IRIRKNLEAN YQENGTKGPL 420 WEEISAGMRR LGYNRSAKRC KEKWENINKY FKKVKESNKK RPLDSKTCPY FHQLEALYNE 480 RNKSGALPML PLMVTPQRQL LLPQETQTEF ETDHRDRAGN KEGEEEGESE EDDYEDEEEE 540 GEGDNETSEF EIVLNKTSSP MDINNNLFT |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 84 | 92 | KRSAKKCKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specific DNA sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00244 | DAP | Transfer from AT1G76890 | Download |
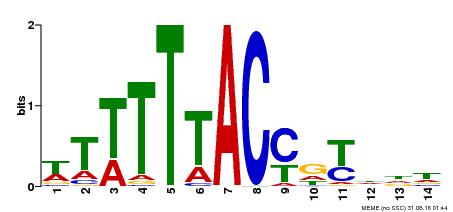
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_01036.1_g00011.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018446613.1 | 0.0 | PREDICTED: trihelix transcription factor GT-2 | ||||
| Swissprot | Q39117 | 0.0 | TGT2_ARATH; Trihelix transcription factor GT-2 | ||||
| TrEMBL | A0A3P6BUA4 | 0.0 | A0A3P6BUA4_BRACM; Uncharacterized protein | ||||
| STRING | Bra015715.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5995 | 28 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76890.2 | 0.0 | Trihelix family protein | ||||



