 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_01129.1_g00005.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 646aa MW: 71867.3 Da PI: 6.1247 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 94.2 | 1.3e-29 | 55 | 140 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+++e+laL+++r+em++++r++ lk+plWe+vs+k+ e g++rs+k+Ckek+en++k++k++ke++++++ + +t+++f+qle
Rsa1.0_01129.1_g00005.1 55 RWPREETLALLRIRSEMDSTFRDATLKAPLWEHVSRKLLELGYKRSAKKCKEKFENVQKYHKRTKETRGGGRH-EGKTYKFFSQLE 139
8*********************************************************************955.5569*******8 PP
trihelix 87 a 87
a
Rsa1.0_01129.1_g00005.1 140 A 140
5 PP
| |||||||
| 2 | trihelix | 103.7 | 1.3e-32 | 408 | 492 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+k+e+laLi++r+ me r++++ k+ lWee+s m++ g++r++k+Ckekwen+nk+ykk+ke++kkr +++ +tcpy+++l+
Rsa1.0_01129.1_g00005.1 408 RWPKEEILALINLRSGMEPRYQDNVPKGLLWEEISTSMKRMGYNRNAKRCKEKWENINKYYKKVKESNKKR-PQDAKTCPYYHRLD 492
8*********************************************************************8.99999*******97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 7.108 | 48 | 112 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.0031 | 52 | 114 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 2.87E-26 | 54 | 119 | No hit | No description |
| Pfam | PF13837 | 2.1E-19 | 54 | 141 | No hit | No description |
| PROSITE profile | PS50090 | 7.329 | 401 | 465 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.0011 | 405 | 467 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.7E-4 | 407 | 464 | IPR009057 | Homeodomain-like |
| Pfam | PF13837 | 7.0E-22 | 407 | 492 | No hit | No description |
| CDD | cd12203 | 2.00E-30 | 408 | 472 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008361 | Biological Process | regulation of cell size | ||||
| GO:0010090 | Biological Process | trichome morphogenesis | ||||
| GO:0030308 | Biological Process | negative regulation of cell growth | ||||
| GO:0032876 | Biological Process | negative regulation of DNA endoreduplication | ||||
| GO:0042631 | Biological Process | cellular response to water deprivation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:2000037 | Biological Process | regulation of stomatal complex patterning | ||||
| GO:2000038 | Biological Process | regulation of stomatal complex development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 646 aa Download sequence Send to blast |
MEQRGGGGGG DEVVEEASPI SSRPPANMEE LMGFSAAVDD GGGLGGSSSS SSGNRWPREE 60 TLALLRIRSE MDSTFRDATL KAPLWEHVSR KLLELGYKRS AKKCKEKFEN VQKYHKRTKE 120 TRGGGRHEGK TYKFFSQLEA LNTTPPPSSL DVAPLSVANP AQQPSPSSSQ FPVFSLTQTP 180 PSQPLPPHTV SFTPNPLPPP PLPSVGPTFP GGTFSSHSSS TASGMESDDE DNMDVDLAGP 240 SSHKRKRENR GGTGPGKMME LFEGLVRQVM QKQAAMQRSF LEAVEKREQE RLAREEAWKR 300 QETSRLAREH EIMSQQRAAS ASRDAAIISL IQKITGHTIQ LPPSLSSQPP HQPPPAAKRP 360 SQAVEPPHLQ PIMAVPQQQV LPPPPPPPPY QQQEMIMSSD QSSPSSSRWP KEEILALINL 420 RSGMEPRYQD NVPKGLLWEE ISTSMKRMGY NRNAKRCKEK WENINKYYKK VKESNKKRPQ 480 DAKTCPYYHR LDLLYSNKVL GSGAGSSTSG LPQDQLSTII QKQSPVSAMK PSQEGDVNVH 540 QTHGSASSDE EEPREETPQG AEKPEDLVMR ELMQQKQHQE DSMIGEYEKI EASHNYNNME 600 IEEEEMEMDE ELDEDEKSTA YEIAFQSPEK RGGNGHTESP FLTMVQ |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 97 | 105 | KRSAKKCKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor that binds specific DNA sequence such as GT3 box 5'-GGTAAA-3' in the SDD1 promoter. Negative regulator of water use efficiency (WUE) via the promotion of stomatal density and distribution by the transcription repression of SDD1. Regulates the expression of several cell cycle genes and endoreduplication, especially in trichomes where it prevents ploidy-dependent plant cell growth. {ECO:0000269|PubMed:19717615, ECO:0000269|PubMed:21169508}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00177 | DAP | Transfer from AT1G33240 | Download |
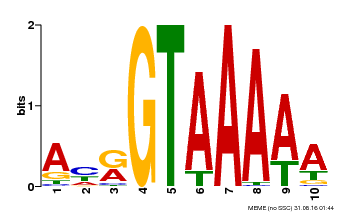
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_01129.1_g00005.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by water stress. {ECO:0000269|PubMed:21169508}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018447113.1 | 0.0 | PREDICTED: trihelix transcription factor GTL1-like isoform X1 | ||||
| Swissprot | Q9C882 | 0.0 | GTL1_ARATH; Trihelix transcription factor GTL1 | ||||
| TrEMBL | A0A397YAF1 | 0.0 | A0A397YAF1_BRACM; Uncharacterized protein | ||||
| TrEMBL | M4F6M1 | 0.0 | M4F6M1_BRARP; Uncharacterized protein | ||||
| STRING | Bra036731.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6226 | 27 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G33240.1 | 1e-137 | GT-2-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



