 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_01415.1_g00004.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 938aa MW: 106843 Da PI: 7.0536 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 161.5 | 1.6e-50 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYah 87
+e +rwl+++ei+a+L n++ +++ ++ + pksg++++++rk++r+frkDG++wkkkkdgkt++E+he+LKvg+ e +++yYah
Rsa1.0_01415.1_g00004.1 30 EEaYTRWLRPNEIHALLSNHNYFTINVKPVHLPKSGTILFFDRKMLRNFRKDGHNWKKKKDGKTIKEAHEHLKVGNEERIHVYYAH 115
55589********************************************************************************* PP
CG-1 88 seenptfqrrcywlLeeelekivlvhylevk 118
+++nptf rrcywlL++++e+ivlvhy+e++
Rsa1.0_01415.1_g00004.1 116 GDDNPTFVRRCYWLLDKSQEHIVLVHYRETH 146
****************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 75.573 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.1E-73 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.2E-45 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.36E-14 | 390 | 476 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.06E-12 | 576 | 686 | No hit | No description |
| Pfam | PF12796 | 1.0E-6 | 576 | 656 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.2E-15 | 577 | 689 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 3.11E-15 | 585 | 692 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 13.761 | 594 | 659 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 2.7E-6 | 627 | 656 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.808 | 627 | 659 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 7.181 | 776 | 802 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 260 | 791 | 813 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.3E-4 | 814 | 836 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.761 | 815 | 839 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 8.7E-5 | 816 | 836 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 11 | 890 | 912 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.334 | 891 | 920 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 938 aa Download sequence Send to blast |
MAGVDSARLI GSEIHGFHTL QDLDIKTMLE EAYTRWLRPN EIHALLSNHN YFTINVKPVH 60 LPKSGTILFF DRKMLRNFRK DGHNWKKKKD GKTIKEAHEH LKVGNEERIH VYYAHGDDNP 120 TFVRRCYWLL DKSQEHIVLV HYRETHEVQA APATPGNSYS SSTTDHLSAK PLAEDNNSGV 180 RNPGEHESPD DLELLQSWLF FCAVHNFYNS LVARNHEISL HEINTLDWDE LLVEADISNQ 240 PSPTEDDVLY FTEQLQTAPM GSAQQGNHHA LYNGSTDIPS YLGLGDPVYQ NNSPCGAREF 300 SSQHLHCVVD PNQQTRDSSA AVANEQGDAL LNNGYGSQES FGKWVHNFIS DSPGSVDDPS 360 LEAVYTPGQE SSAPPAVFHP QSNIPDQIFN ITDVSPAWAY STEKTKILVT GFFHDSFQHF 420 GRSNLFCICG EVRVPAEFLQ MGVYRCFVPP QSPGIVNLYL SADGSKPISQ LFSFEHRSVP 480 VIEKVVPQED QLYKWEEFEF QVRLAHLLFT SSNKISVFSS KISPDNLLEA KKLASRTSHL 540 LNSWAYLMKS IQANEVPFDQ ARDHLFELTL KNRLKEWLLE KVIEDRNTKE YDSKGLGVIH 600 LCAVLGYTWS ILLFSWANIS LDFRDKHGWT ALHWAAYYGR EKMVAALLSA GARPNLVTDP 660 TKEYLGGCTA ADIAQQKGYE GLAAFLAEKC LVAQFRDMKM AGNISGNLEG IKAETSANPG 720 HSNEEEQSLK DTLAAYRTAA EAAARIQGAF REHELKVRSK AVRFASKEEE AKNIIAAMKI 780 QHAFRNYETR RKIAAAARIQ YRFQTWKMRR EFLNMRKKAI KIQAVFRGFQ VRRQYQKITW 840 SVGVLEKAIL RWRLKRRGFR GLQVSQPEAM EGTDVVEDFY KTSQKQAEDR LERSVVRVQA 900 MFRSKKAQQD YRRMKLAHEE AQLEYDGMQE LDHMAMES |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
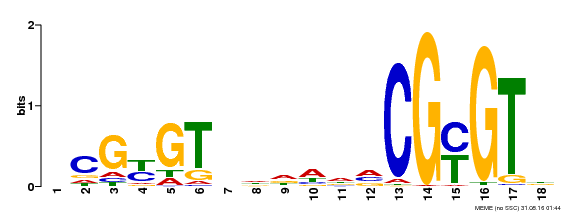
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_01415.1_g00004.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY128295 | 0.0 | AY128295.1 Arabidopsis thaliana AT4g16150/dl4115w mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018450972.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A3P6GMJ4 | 0.0 | A0A3P6GMJ4_BRAOL; Uncharacterized protein | ||||
| STRING | Bo8g043280.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4021 | 27 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



