 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_01719.1_g00001.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 982aa MW: 110666 Da PI: 5.9008 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 185.6 | 5.2e-58 | 5 | 122 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYa 86
l+e ++rwl++ ei++iL n++k+++++e++trp sgsl+L++rk++ryfrkDG++w+kkkdgkt++E+hekLKvg+++vl+cyYa
Rsa1.0_01719.1_g00001.1 5 LSEaQHRWLRPAEICEILRNYHKFHIATESPTRPASGSLFLFDRKVLRYFRKDGHNWRKKKDGKTIKEAHEKLKVGSIDVLHCYYA 90
45559********************************************************************************* PP
CG-1 87 hseenptfqrrcywlLeeelekivlvhylevk 118
h+e n++fqrrcyw+Le+el +iv+vhylevk
Rsa1.0_01719.1_g00001.1 91 HGEGNENFQRRCYWMLEQELMHIVFVHYLEVK 122
*****************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 84.274 | 1 | 127 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.2E-84 | 4 | 122 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.0E-50 | 7 | 121 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 6.12E-14 | 391 | 477 | IPR014756 | Immunoglobulin E-set |
| PROSITE profile | PS50297 | 18.643 | 574 | 656 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.55E-17 | 575 | 685 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.9E-16 | 576 | 686 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 3.54E-12 | 580 | 683 | No hit | No description |
| Pfam | PF12796 | 9.7E-7 | 581 | 651 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.0029 | 624 | 653 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.22 | 624 | 656 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 1.9 | 800 | 822 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 1.01E-7 | 800 | 850 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.693 | 801 | 830 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0012 | 802 | 821 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0029 | 823 | 845 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.322 | 824 | 848 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.1E-4 | 826 | 845 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 982 aa Download sequence Send to blast |
MEQLLSEAQH RWLRPAEICE ILRNYHKFHI ATESPTRPAS GSLFLFDRKV LRYFRKDGHN 60 WRKKKDGKTI KEAHEKLKVG SIDVLHCYYA HGEGNENFQR RCYWMLEQEL MHIVFVHYLE 120 VKGSRTSIGM KENNSNSLSG SVSVNIDSAA SPTSRLSSYC EDADSVFGGA GDSHQSSSVL 180 QASPEPQTGN HNGWTSAPGM RSDSQRLFDV QAWDAVGNLV TRYDQPCNNL LVEERTDKGG 240 MLAAEHLRSP LQTQLNWQIP AQDDLPLPKW HLAPHSGMAD DTDLALFEQS AQDNFESFSS 300 LLDIEHLQSD GIPPSNMETE FIPVKKSLLR HEDSLKKVDS FSRWASKELG EMEDLQMQSS 360 RGDIAWTTVD CETAAAGLAF SPSLSEDQRF TIVDYWPKCA QTDAEVEVLV IGTFLLSPQE 420 VTICSWSCMF GEVEVPAEIL VDGVLCCHAP PHTAGQVPFY VTCSNRFACS ELREFEFLSG 480 STKKIDAADI YGYSSKETSL QLRFEKLLAH RDFVQEHQIF EDVVEKRRKI SSIMLLNEEK 540 EYLFPGMYER DSTKQEPKER VFREQFEDEL YIWLIHKVTE GGKGPNLLDE GGQGVLHFVA 600 ALGYDWAIKP ILAAGVNINF RDSNGWSALH WAAFSGREET VAVLVSLGAD AGALTDPSPE 660 LPLGKTAADL AYGKEHRGIS GFLAESSLTS YLEKLTMVSK ENSPANSSGS KAVQTVSERT 720 AAPMSYGDVP EKLSMKDSLT AVRNATQAAN RLHQVFRMQS FQRKQLSGFD DDDDELGISN 780 ELAVSFAASK TKNPGQSEVF AHSAATHIQK KYRGWKKRKE FLLIRQRVVK IQAHVRGHQV 840 RKQYKPIVWS VGLLEKIILR WRRKGTGLRG FKRNVVPKTV EPEPLSPMIP KEDDYDFLEK 900 GRKQTEERLQ KALTRVKSMV QYPEARDQYR RLLTVVEGFR ENEASSSLSI NNREEGLVNC 960 EEDDLIDIES ILNDDILMST SP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro (PubMed:11925432). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance (PubMed:19270186). Involved in freezing tolerance in association with CAMTA2 and CAMTA3. Contributes together with CAMTA2 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved in drought stress responses by regulating several drought-responsive genes (PubMed:23547968). Involved in auxin signaling and responses to abiotic stresses (PubMed:20383645). Activates the expression of the V-PPase proton pump AVP1 in pollen (PubMed:14581622). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:11925432, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:23547968, ECO:0000269|PubMed:23581962, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
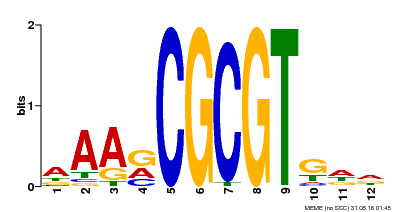
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_01719.1_g00001.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by UVB, wounding, ethylene and methyl jasmonate (PubMed:12218065). Induced by salt stress and heat shock (PubMed:12218065, PubMed:20383645). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF491304 | 0.0 | AF491304.1 Brassica napus calmodulin-binding transcription activator (CAMTA) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018446094.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 1 isoform X2 | ||||
| Refseq | XP_018446095.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 1 isoform X2 | ||||
| Swissprot | Q9FY74 | 0.0 | CMTA1_ARATH; Calmodulin-binding transcription activator 1 | ||||
| TrEMBL | M4CYT5 | 0.0 | M4CYT5_BRARP; Uncharacterized protein | ||||
| STRING | Bra009382.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||



