 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Rsa1.0_02923.1_g00001.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Raphanus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 243aa MW: 28125.7 Da PI: 9.7189 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.5 | 6.2e-18 | 5 | 50 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg W + Ede+l ++v+q+G+++W++Ia+++ gR++k+c++rw++
Rsa1.0_02923.1_g00001.1 5 RGHWRPAEDEKLRELVQQFGPHNWNAIAQKLS-GRSGKSCRLRWFNQ 50
899*****************************.***********996 PP
| |||||||
| 2 | Myb_DNA-binding | 55.6 | 1.3e-17 | 57 | 100 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
r+++T+eE+e+l+ ++ +G++ W+ Iar ++ gRt++ +k++w+
Rsa1.0_02923.1_g00001.1 57 RNPFTEEEEERLLASHRIHGNR-WSVIARFFP-GRTDNAVKNHWHV 100
789*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 24.726 | 1 | 55 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.4E-14 | 4 | 53 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.8E-17 | 5 | 50 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 4.12E-30 | 5 | 98 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-27 | 6 | 58 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.41E-13 | 8 | 49 | No hit | No description |
| PROSITE profile | PS51294 | 18.713 | 56 | 106 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.3E-15 | 56 | 104 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.7E-14 | 57 | 99 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.91E-12 | 59 | 99 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 6.7E-21 | 59 | 104 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 243 aa Download sequence Send to blast |
MMCSRGHWRP AEDEKLRELV QQFGPHNWNA IAQKLSGRSG KSCRLRWFNQ LDPRINRNPF 60 TEEEEERLLA SHRIHGNRWS VIARFFPGRT DNAVKNHWHV IMARRGRERS KVRPRSLGHD 120 GTAVGPGMMG YKDTDKKRRL GTTTAINYPY SFSHINHFQI LKEFLTGKIG FCNNTTPINE 180 GAVDQTKRPL EFYNFLQVKT DSKKPEVIHN SRKDEEEEED VDSCVPFFDF LSVGNSASHQ 240 GLC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 1e-31 | 2 | 105 | 1 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 1e-31 | 2 | 105 | 1 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that confers sensitivity to abscisic acid (ABA) and salt, but tolerance to drought (PubMed:21399993). Regulates secondary cell wall (SCW) biosynthesis, especially in interfascicular and xylary fibers (PubMed:18952777, PubMed:23781226). {ECO:0000269|PubMed:18952777, ECO:0000269|PubMed:21399993, ECO:0000269|PubMed:23781226}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00145 | DAP | Transfer from AT1G17950 | Download |
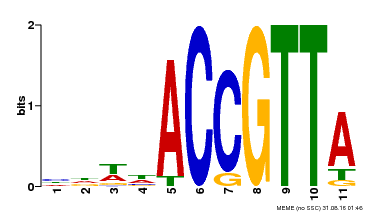
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Rsa1.0_02923.1_g00001.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (PubMed:16463103, PubMed:21399993). Accumulates in response to salt (PubMed:21399993). Triggered by MYB46 and MYB83 in the regulation of secondary cell wall biosynthesis (PubMed:19674407, PubMed:22197883). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:19674407, ECO:0000269|PubMed:21399993, ECO:0000269|PubMed:22197883}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY519554 | 0.0 | AY519554.1 Arabidopsis thaliana MYB transcription factor (At1g17950) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018449713.1 | 1e-178 | PREDICTED: transcriptional activator Myb-like isoform X1 | ||||
| Swissprot | Q6R0C4 | 1e-157 | MYB52_ARATH; Transcription factor MYB52 | ||||
| TrEMBL | A0A0D3DQ45 | 1e-162 | A0A0D3DQ45_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3P6GAB3 | 1e-162 | A0A3P6GAB3_BRAOL; Uncharacterized protein | ||||
| STRING | Bo8g067590.1 | 1e-163 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2818 | 27 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G17950.1 | 1e-154 | myb domain protein 52 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



