 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00000662-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 267aa MW: 30007.1 Da PI: 10.0669 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 104.9 | 4.8e-33 | 64 | 118 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprlrWt +LH+rFv+av++LGG++kAtPk+++++m++kgLtl+h+kSHLQkYRl
SMil_00000662-RA_Salv 64 KPRLRWTADLHDRFVDAVTKLGGPHKATPKSVMRVMGLKGLTLYHLKSHLQKYRL 118
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-31 | 61 | 121 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.649 | 61 | 121 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.16E-15 | 64 | 121 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 6.7E-24 | 64 | 119 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.1E-8 | 66 | 117 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 1.1E-22 | 157 | 202 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 267 aa Download sequence Send to blast |
MAVPSLKDVA RDHRNIRVLT SAGGDSALPY TCPHIYTVHR SHRRTFPTPR VGGSGGVVLT 60 RDPKPRLRWT ADLHDRFVDA VTKLGGPHKA TPKSVMRVMG LKGLTLYHLK SHLQKYRLGQ 120 QHAKQHTNTD NFENPYMHIA STSTNSSSMN SEQGEIPVAE AVRCQIEVQK TLEEQLEVQK 180 KLQMRIEAQG KYLQAILERA HRSLSESAKA QLTTDFNLAL SSFMHTVNEE PRDTKLVLKL 240 EGPSIDFDLN SRSSYDFIGT SKSMNLH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 7e-19 | 64 | 117 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 7e-19 | 64 | 117 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 7e-19 | 64 | 117 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 7e-19 | 64 | 117 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00544 | DAP | Transfer from AT5G45580 | Download |
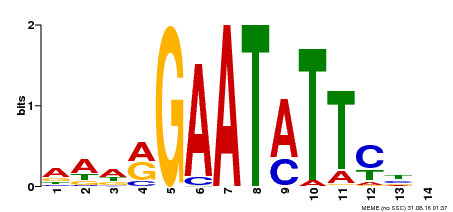
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012854303.1 | 1e-101 | PREDICTED: myb family transcription factor APL | ||||
| Swissprot | C0SVS4 | 1e-69 | PHLB_ARATH; Myb family transcription factor PHL11 | ||||
| TrEMBL | A0A4D9BJZ5 | 1e-123 | A0A4D9BJZ5_SALSN; Uncharacterized protein | ||||
| STRING | Migut.A00576.1.p | 1e-100 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4416 | 21 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45580.1 | 4e-66 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



