 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00000992-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 501aa MW: 55576.3 Da PI: 5.6508 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 36.7 | 7.5e-12 | 355 | 401 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L + YI +Lq
SMil_00000992-RA_Salv 355 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDSIAYITDLQ 401
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 3.4E-38 | 49 | 228 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.387 | 351 | 400 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.04E-13 | 354 | 405 | No hit | No description |
| SuperFamily | SSF47459 | 2.09E-17 | 354 | 420 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.3E-9 | 355 | 401 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 9.5E-17 | 355 | 412 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.1E-14 | 357 | 406 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 501 aa Download sequence Send to blast |
MGKKVWWNEE EKVVVESVLG SEATEYFDWA ASNNVLSEFS TSSGDLGVQE ALCKIVEGSD 60 LTYAIYWHVS KSKSGRSALI WGDGHYQESK ESVHDSCGSY NDLKRVEGDR RKWVLQKLHA 120 CFGGLEDDNV AAKLDQVSNV EMLYLTSMFF VFPFDKPSIP SQSFNSDRCI WVSNLDSCLE 180 RYHSRAYLAK LAQFETVAFV PTKSGVVEIG SRKSIPENKS IIKAAKSIVV AFNSTQVKAV 240 PKIFGQDLSM GGSRSGTINI SFSPKVEDDS GFSAESYDLQ AISSNHGYAN SSNGHCGDDG 300 DTKVFSHHVV AAGLDSQSII SGMELGKEDC FFLSDDQRPR KRGRKPANGR EEPLNHVEAE 360 RQRREKLNQR FYALRAVVPN ISKMDKASLL GDSIAYITDL QAKIRVLEAE KGIGNTNQQL 420 NPIPDFEFHE RLEDAVLRVS YTLETHPVSK VVKALREQQV MAQESSISID DSGEVVHTFS 480 IQTESGDAEQ LKDKLTAALL K |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 6e-29 | 349 | 411 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 6e-29 | 349 | 411 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 6e-29 | 349 | 411 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 6e-29 | 349 | 411 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 6e-29 | 349 | 411 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 6e-29 | 349 | 411 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 6e-29 | 349 | 411 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 6e-29 | 349 | 411 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that negatively regulates jasmonate (JA) signaling (PubMed:30610166). Negatively regulates JA-dependent response to wounding, JA-induced expression of defense genes, JA-dependent responses against herbivorous insects, and JA-dependent resistance against Botrytis cinerea infection (PubMed:30610166). Plays a positive role in resistance against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PubMed:30610166). {ECO:0000269|PubMed:30610166}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00099 | PBM | Transfer from AT4G16430 | Download |
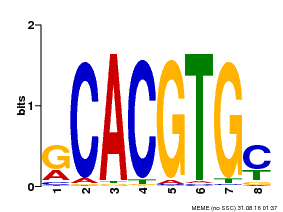
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding, feeding with herbivorous insects, infection with the fungal pathogen Botrytis cinerea and infection with the bacterial pathogen Pseudomonas syringae pv tomato DC3000. {ECO:0000269|PubMed:30610166}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KP257470 | 0.0 | KP257470.1 Salvia miltiorrhiza clone 37 basic helix-loop-helix transcription factor mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011080018.1 | 0.0 | transcription factor bHLH3-like | ||||
| Refseq | XP_011080019.1 | 0.0 | transcription factor bHLH3-like | ||||
| Swissprot | A0A3Q7H216 | 0.0 | MTB3_SOLLC; Transcription factor MTB3 | ||||
| TrEMBL | A0A0H3YC31 | 0.0 | A0A0H3YC31_SALMI; Basic helix-loop-helix transcription factor | ||||
| STRING | Migut.D00401.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA6669 | 24 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16430.1 | 1e-141 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



