 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00001046-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 378aa MW: 40367.7 Da PI: 9.3393 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 47.9 | 2.4e-15 | 215 | 260 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn+ Er+RRd+iN+++ +L++l+P++ K +Ka++L +++eY+k+Lq
SMil_00001046-RA_Salv 215 HNQSERKRRDKINQRMKTLQKLVPNS-----SKSDKASMLDEVIEYLKQLQ 260
9************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 17.504 | 210 | 259 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 6.54E-19 | 213 | 283 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.7E-18 | 215 | 268 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.84E-17 | 215 | 264 | No hit | No description |
| Pfam | PF00010 | 5.8E-13 | 215 | 260 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.2E-17 | 216 | 265 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 378 aa Download sequence Send to blast |
MGEWSARHAW PGVPRVVNKP SASSPPSTWD KPRASGTLES IVNQATSLGE KESELGGHRA 60 KANPAAVAMD ALVPCGGGGH EQQHPAAAQA HVRGASRAGC STRVSKCSGA RVGSRSVGES 120 ATCGGGSDSR QTISIDTWGE REFGGPGFTS TASLISPENT ISGQDYTKTS ADDRDSVTLH 180 SGSQASHRKG DEEGRRRDNG KSSVCTKRSR AAAIHNQSER KRRDKINQRM KTLQKLVPNS 240 SKSDKASMLD EVIEYLKQLQ AQVQMMSRMN MPSMMLPLAM QQQQFQMSMM GCMGMGMGMG 300 MGVMDVPRPP FMPVPAAAAA WDNGGDCLPP PPPPVAMPDP LAALLACQSQ PMTMDAYSRV 360 AAMYQQLQQQ QQLPASKN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 218 | 223 | ERKRRD |
| 2 | 219 | 224 | RKRRDK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Required during the fertilization of ovules by pollen. {ECO:0000269|PubMed:15634699}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
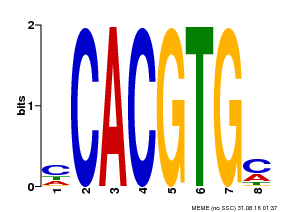
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KP257472 | 0.0 | KP257472.1 Salvia miltiorrhiza clone 39 basic helix-loop-helix transcription factor mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011093295.1 | 1e-112 | LOW QUALITY PROTEIN: transcription factor UNE10 | ||||
| Swissprot | Q8GZ38 | 3e-63 | UNE10_ARATH; Transcription factor UNE10 | ||||
| TrEMBL | A0A0H3YB30 | 0.0 | A0A0H3YB30_SALMI; Basic helix-loop-helix transcription factor | ||||
| STRING | cassava4.1_007418m | 7e-98 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4367 | 21 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 2e-48 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



