 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00001178-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 506aa MW: 55988.6 Da PI: 9.4933 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 92 | 5.1e-29 | 276 | 329 | 1 | 54 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYR 54
k+r++W+p+LH+rFv+a++ LGGs++AtPk+i+elmkv+gLt+++vkSHLQ Y
SMil_00001178-RA_Salv 276 KARRCWSPDLHRRFVNALHMLGGSQVATPKQIRELMKVDGLTNDEVKSHLQTYN 329
68***************************************************6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.273 | 273 | 333 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.55E-15 | 273 | 334 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 7.2E-25 | 274 | 333 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.1E-23 | 276 | 329 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.2E-6 | 278 | 328 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009266 | Biological Process | response to temperature stimulus | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010452 | Biological Process | histone H3-K36 methylation | ||||
| GO:0048579 | Biological Process | negative regulation of long-day photoperiodism, flowering | ||||
| GO:1903507 | Biological Process | negative regulation of nucleic acid-templated transcription | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 506 aa Download sequence Send to blast |
LQSPNLIDKN ESAYRVFLVL DIPLHDSTVI PSTRSSHTES KEDCLIYVID QEMMASPSEL 60 SFDCKPQIYS MLPKSFGDQA SEHTQKLEEF LGRLEEERLK IDAFKRELPL CMQLLSNAME 120 ASRQQLQSER ASQGGRPVLE EFIPLKTSNS SSVEIGEGNM SADKANWMTS AQLWNQESEG 180 AKAPQSPLTS PQETDIKLAL NCKQRVNGGG AFLPFSSKDR ASSPSGEAYP ELALASSHKE 240 EKRSSSETKR ESAGKCSVEK EQACGTATNG SQTHRKARRC WSPDLHRRFV NALHMLGGSQ 300 VATPKQIREL MKVDGLTNDE VKSHLQTYNL WFKNKTKTLK FVKAEIPTSH SKAQPEPTSG 360 GHGGPSGGGP RWHMGPARIR RRRRRTRRGT GPLRSAPRHH PPRVAALLLP KPADAARHVP 420 RHSSPPNHQL HHHQQLHFYN KQPPVTQRNS SPESDVRGGG GDPSESIEDG KSESGSWKAD 480 SGGDNSGERR RVEGEESNAS VVSLKF |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 377 | 384 | RIRRRRRR |
| 2 | 379 | 386 | RRRRRRTR |
| 3 | 379 | 387 | RRRRRRTRR |
| 4 | 380 | 385 | RRRRRT |
| 5 | 380 | 387 | RRRRRTRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor acting as a flowering repressor, directly repressing FT expression in a dosage-dependent manner in the leaf vasculature. {ECO:0000269|PubMed:25132385}. | |||||
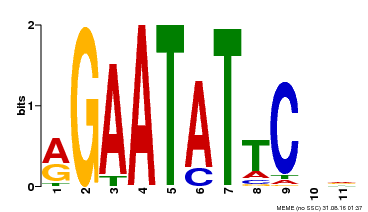
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00261 | DAP | Transfer from AT2G03500 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by SVP. Down-regulated by high temperature and gibberellic acid treatment. Not regulated by photoperiod, circadian rhythm under long days or vernalization. {ECO:0000269|PubMed:25132385}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028753830.1 | 1e-128 | myb family transcription factor EFM-like | ||||
| Refseq | XP_028800631.1 | 1e-128 | myb family transcription factor EFM-like | ||||
| Swissprot | Q9ZQ85 | 5e-82 | EFM_ARATH; Myb family transcription factor EFM | ||||
| TrEMBL | A0A4D8XRP1 | 1e-153 | A0A4D8XRP1_SALSN; Uncharacterized protein | ||||
| STRING | evm.model.supercontig_127.55 | 1e-125 | (Carica papaya) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA8794 | 22 | 29 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03500.1 | 3e-73 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



