 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00001418-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 508aa MW: 56370.2 Da PI: 5.1365 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 116.5 | 1.6e-36 | 18 | 110 | 2 | 103 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkg 89
Fl+k+y++++d++++ ++sws+n+nsfvv++ efa+++LpkyFkh+nf+SFvRQLn+YgF+kv+ ++ weF+++ F +g
SMil_00001418-RA_Salv 18 FLSKTYDMVDDPATDAVVSWSKNNNSFVVWNVPEFARDLLPKYFKHNNFSSFVRQLNTYGFRKVDPDR---------WEFANEGFLRG 96
9****************************************************************999.........*********** PP
XXXXXXXXXXXXXX CS
HSF_DNA-bind 90 kkellekikrkkse 103
+k+ll++i+r+k++
SMil_00001418-RA_Salv 97 RKHLLKTINRRKTS 110
***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 3.7E-39 | 11 | 102 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.36E-35 | 13 | 107 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 2.6E-62 | 14 | 107 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 7.1E-32 | 18 | 107 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 2.5E-20 | 18 | 41 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 2.5E-20 | 56 | 68 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 57 | 81 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 2.5E-20 | 69 | 81 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009408 | Biological Process | response to heat | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 508 aa Download sequence Send to blast |
MEAGGGGIST AATSPPPFLS KTYDMVDDPA TDAVVSWSKN NNSFVVWNVP EFARDLLPKY 60 FKHNNFSSFV RQLNTYGFRK VDPDRWEFAN EGFLRGRKHL LKTINRRKTS HAQGQQQNVQ 120 VQNPSVPSCI EVGKFGIEEE VERLKRDKNL LMQELVRLRQ QQQSTDSQLQ TVGQRVHVME 180 QRQTQMMSFL AKAMQSPGFV SQLVHQQNDS GRHISGANKK RRLPNQDEES LARKLSITSP 240 DGQIVKYQPL MNEAAKAMLR QIMKMNTSNR PESKLSNPNG FLIDSAHPPS DLIDSSSSSS 300 RISGVTLSEV LPNTSTLSEV QSSSCLTHAS AHNPLFPEAS TGLPDFIQAQ GISPGNSIDP 360 PNGPFKGSEP TSFSSVDALP GFVDAPIPPP SDEPLEDYDV DILLDDIHKL AAMDDDIHKL 420 SSMSDDIHKM PGISDDIHKL PGINDVFWEQ FLPESPLIEK TDEISTVDLG HEMSTDQLIE 480 LESEWDRLKS LNNLTEQMGL LASSAKIG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ldu_A | 9e-27 | 16 | 110 | 18 | 123 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds DNA of heat shock promoter elements (HSE). {ECO:0000250}. | |||||
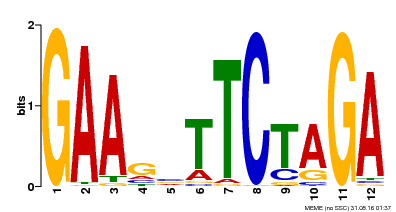
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00514 | DAP | Transfer from AT5G16820 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Not induced by heat stress. {ECO:0000269|PubMed:18064488}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011084620.1 | 0.0 | heat stress transcription factor A-1-like | ||||
| Swissprot | Q84T61 | 1e-157 | HSFA1_ORYSJ; Heat stress transcription factor A-1 | ||||
| TrEMBL | A0A4D9A0V0 | 0.0 | A0A4D9A0V0_SALSN; Heat shock transcription factor, other eukaryote | ||||
| STRING | Migut.F00088.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA5054 | 21 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G16820.2 | 1e-132 | heat shock factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



