 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00006890-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 772aa MW: 83819.2 Da PI: 6.7559 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.6 | 3.5e-16 | 489 | 535 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
SMil_00006890-RA_Salv 489 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 535
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 1.98E-17 | 479 | 539 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.9E-20 | 482 | 544 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.7E-20 | 482 | 546 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 485 | 534 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.2E-13 | 489 | 535 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-17 | 491 | 540 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 772 aa Download sequence Send to blast |
LKIDTGKHPE KRLYLLGDVK FLADFIWSLY WWDFFQPWTF FNSIKLLEFQ RKFCFSTMPL 60 SEFLRMARGK LESGHQKPAP ADISSRPGNE LVELVWENGQ VVMQGQSSRA TRSPTLNNLL 120 SNTSKARDAA TTARYGKFGG VDSIMNDMVP VVPSGDIDLG QDDEIAPWLS YPIDDALGQD 180 YASEILPHIS SVTANGSPAQ NSFVSADKRS SCEQAVNNLQ TGASNVKVPS RSSSKDRLFG 240 SWLPQQHRQT SDALGSGVTD NSSDHLDSVF GNPGQSRDTV NGSASTVMDR RSIPPPANCS 300 NFLNFSHFSR PAALAKGSLS NSDGIPMSVS SVVERVETKD KANCSNPVKS IRIEQVKSMP 360 KDNDSHGSCT LPVVGSREQV IKEPLERADN LFKETSIKND KPLILSNNES SAKGAPDGER 420 IVEPMVASSS VGSGNSADRV SCEQTQHSKR KFCDIEESEC RSDDVETESV DAKKATCLRG 480 SKRSRAAEVH NLSERRRRDR INEKMRALQE LIPNCNKVDK ASMLDEAIEY LKTLQLQVQI 540 MSMSSGLCMP PPMMFPTGMQ HMHPAHVPHF QPMGVGVGMG MAFGMGMPDM NGGSSGCPMY 600 PVPPLQVPHF SSPMLGLANF QRMPGHNHPV FGHPGQAFPN PVTKPPFVPL APRPPVTSAM 660 GSGALRNGSN GEILSTSQIM KSGDPVTTTN SQSMCSAEAR SSVHNKSNQP TKEVVDQCAA 720 VQDNERATDA GSSAAHLSAT TTDINKEPGG VEVLAEWHSP VDDVFLCSPG RT |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 493 | 498 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
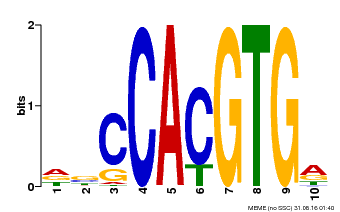
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KP257505 | 0.0 | KP257505.1 Salvia miltiorrhiza clone 72 basic helix-loop-helix transcription factor mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020552005.1 | 0.0 | transcription factor PIF3 isoform X2 | ||||
| TrEMBL | A0A0H3YC53 | 0.0 | A0A0H3YC53_SALMI; Basic helix-loop-helix transcription factor | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4965 | 24 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 2e-61 | phytochrome interacting factor 3 | ||||



