 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00010393-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 406aa MW: 44693 Da PI: 9.0261 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 54.9 | 2.1e-17 | 111 | 160 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
SMil_00010393-RA_Salv 111 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTALAAARAYDRAAIKFRG 160
78*******************......55************************997 PP
| |||||||
| 2 | AP2 | 42 | 2.3e-13 | 203 | 253 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+++GV+ +k grW+A+ + +k+++lg f+t+ eAa+a+++a+ k +g
SMil_00010393-RA_Salv 203 SKFRGVTLHK-CGRWEARMGQF--L-GKKYIYLGLFDTEVEAARAYDKAAIKCNG 253
79********.7******5553..2.26**********99**********99776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 4.6E-9 | 111 | 160 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.48E-15 | 111 | 168 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 5.14E-11 | 112 | 167 | No hit | No description |
| SMART | SM00380 | 2.0E-30 | 112 | 174 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 9.3E-17 | 112 | 168 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.477 | 112 | 168 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.44E-25 | 203 | 261 | No hit | No description |
| Pfam | PF00847 | 1.1E-8 | 203 | 253 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 9.81E-17 | 203 | 261 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 15.185 | 204 | 261 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.9E-32 | 204 | 267 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 5.6E-16 | 204 | 261 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-5 | 205 | 216 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-5 | 243 | 263 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 406 aa Download sequence Send to blast |
MMFDLNLSVG CNGRQSPATS VSSVVDGAGE TSSSNDDASS SVALNFGIIG ADRDRDRAAL 60 VTRELFPASR LGELSDPVIL GAPTRTVMPA QQQVQQVQAR KSRRGPRSRS SQYRGVTFYR 120 RTGRWESHIW DCGKQVYLGG FDTALAAARA YDRAAIKFRG VDADINFNLC DYEEDMKHTK 180 NMSKEEFVHI LRRQSTGFSR GSSKFRGVTL HKCGRWEARM GQFLGKKYIY LGLFDTEVEA 240 ARAYDKAAIK CNGREAVTNF DSTSYDGEMM PHSCSGNDLD LNLGISTSSP KETANLGSFQ 300 FQNANMFKLH VENSAATKPC DSPLNGLPVI LNPPNYEERA GQGRTGLGST SEEVVKWAWR 360 MHDQVRATPI IAASSGFSSP YPTALNFCFS SNPTNVSHIR SSSPRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition. Together with SNB, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes. Prevents lemma and palea elongation as well as grain growth (By similarity). {ECO:0000250|UniProtKB:P47927, ECO:0000250|UniProtKB:Q84TB5}. | |||||
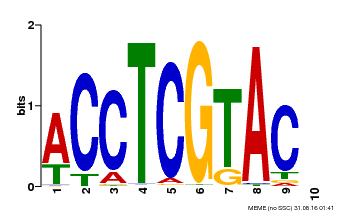
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ784462 | 0.0 | KJ784462.1 Salvia miltiorrhiza AP11/EREBP Transcription Factors (WRI11) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011089852.1 | 1e-160 | ethylene-responsive transcription factor RAP2-7 isoform X1 | ||||
| Swissprot | B8AMA9 | 1e-101 | AP22_ORYSI; APETALA2-like protein 2 | ||||
| TrEMBL | A0A0G2SJB9 | 1e-162 | A0A0G2SJB9_SALMI; AP11/EREBP Transcription Factors (Fragment) | ||||
| STRING | PGSC0003DMT400065313 | 1e-121 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA591 | 24 | 116 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 6e-99 | related to AP2.7 | ||||



