 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | SMil_00018832-RA_Salv | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lamiaceae; Nepetoideae; Mentheae; Salvia
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 376aa MW: 43844 Da PI: 8.8457 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 17.6 | 1.1e-05 | 45 | 69 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ C+ dC s++rk++L+rH+ H
SMil_00018832-RA_Salv 45 FICSvdDCESSYRRKDHLNRHMLQH 69
789999****************988 PP
| |||||||
| 2 | zf-C2H2 | 13.4 | 0.00023 | 74 | 99 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++Cp C+++F+ + n++rH + H
SMil_00018832-RA_Salv 74 FECPveGCKTMFTLQGNMTRHVKDfH 99
89*******************98766 PP
| |||||||
| 3 | zf-C2H2 | 12.9 | 0.00034 | 130 | 154 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
+Cp dC+++F+ + ++ H + H
SMil_00018832-RA_Salv 130 ECPvkDCKRTFKLQGYMAWHVKEyH 154
69999****************9888 PP
| |||||||
| 4 | zf-C2H2 | 15.7 | 4.1e-05 | 168 | 192 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C +Cgk+F+ +s L +H +H
SMil_00018832-RA_Salv 168 HVCAetHCGKVFRYPSKLYKHEDSH 192
789999***************9888 PP
| |||||||
| 5 | zf-C2H2 | 21.4 | 6.6e-07 | 259 | 283 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
+C+ C+ +FsrksnL++H++ H
SMil_00018832-RA_Salv 259 SCSvkGCSLTFSRKSNLNKHMKAvH 283
588889***************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 4.16E-7 | 42 | 71 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 6.8E-10 | 42 | 71 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.743 | 45 | 74 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0026 | 45 | 69 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 47 | 69 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 9.16E-5 | 72 | 99 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 5.2E-5 | 72 | 99 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.014 | 74 | 104 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.012 | 74 | 99 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 76 | 99 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.088 | 129 | 154 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 131 | 154 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 1.7 | 168 | 192 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.242 | 168 | 197 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 170 | 192 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 3 | 200 | 225 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 202 | 225 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 29 | 228 | 249 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 2.7E-6 | 239 | 280 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.08E-9 | 240 | 299 | No hit | No description |
| PROSITE profile | PS50157 | 12.05 | 258 | 288 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 3.2E-4 | 258 | 283 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 260 | 283 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-5 | 287 | 311 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 12 | 289 | 315 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 291 | 315 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 376 aa Download sequence Send to blast |
LDPNKKNSKM REATNLDWIS TRWVTAWEAP SSVPRSRLLF LERPFICSVD DCESSYRRKD 60 HLNRHMLQHQ GKLFECPVEG CKTMFTLQGN MTRHVKDFHC ESAPTDIEHP KESNCGKLHK 120 HEDSQEKLLE CPVKDCKRTF KLQGYMAWHV KEYHGESAFT DVKHTTEHVC AETHCGKVFR 180 YPSKLYKHED SHVKLDSLEA YCAEPGCMKY FSNERCLKEH IRSCHQYVLC DECGNKQLKK 240 NIKRHMRMHE AEHSSGRISC SVKGCSLTFS RKSNLNKHMK AVHLKLKPFA CGMPGCDKRF 300 TFKHVRNNHE KSSCHVYTPG GDFEASDEQF RSRPRGGRKR KYPVIETLMR KRIVPPSESE 360 LAMNEGQDYP SWQFSN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 332 | 340 | RPRGGRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
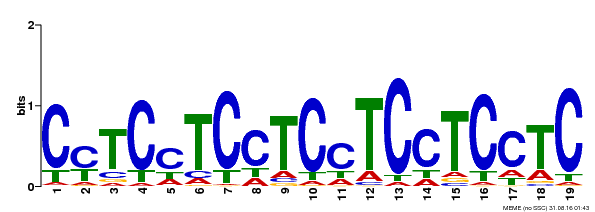
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011079244.1 | 1e-136 | transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 1e-111 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A4D8Z9A3 | 0.0 | A0A4D8Z9A3_SALSN; Uncharacterized protein | ||||
| STRING | Migut.K00168.1.p | 1e-121 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3644 | 24 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.1 | 1e-114 | transcription factor IIIA | ||||



